Introduction
The insulin receptor is a transmembrane receptor [1] that resides in the plasma membrane and is activated by the binding of insulin. The insulin receptor belongs to the large class of receptor tyrosine kinase (RTKs). RTKs are found at the cell surface and have a high affinity for a particular ligand. RTKs are made up of three distinct parts: an extracellular domain with ligand binding sites, a transmembrane region, and an intracellular domain with the tyrosine kinases that initiate intracellular signaling cascades [1]. Downstream signaling from the insulin receptor initiates a variety of cellular pathways including glucose homeostasis, regulation of lipid, protein, and carbohydrate metabolism, gene expression, and even modulation of brain neurotransmitter levels [2]. Amongst RTKs, the insulin receptor is unusual as its undergoes a large conformation change upon insulin binding. Through recent cryo-EM structures of the insulin receptor bound in various conformations, a complete three-dimensional understanding of this conformational changes in finally coming into focus. This page focuses specifically on the insulin receptor's role in glucose homeostasis.
Structural Overview
The insulin receptor is a dimer of heterodimers made of two and two [2]. Within the extracellular ectodomain, there are four potential that can interact with insulin ligands on the extracellular side of the membrane. The insulin receptor has only been imaged in pieces, which has led to proposed structures of the entirety of the receptor based on the function of downstream tyrosine autophosphorylation.PDB 1IR3
Alpha Subunits
The alpha subunits make up the extracellular domain (ectodomain) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. The alpha subunits are held together by a disulfide bond between at the CYS524 position on each alpha subunit. Two types of insulin binding sites are present in the alpha subunits, and . The sites are in pairs because of the heterodimeric nature of the receptor. Due to structural differences, as well as greater surface area and accessibility, binding sites 1 and 1' have much higher affinity than that of sites 2 and 2'. Insulin can also bind at sites 2 and 2', but the location on the back of the beta sheet of the FnIII-1 domain and lack of surface area decreases the likelihood of their binding site becoming occupied as quickly. [3] Cryo-EM has imaged insulin bound structures that displayed a T-shape conformation in the alpha subunits.[3]
Beta Subunits
The beta subunit spans from the extracellular domain across the transmembrane region and into the intracellular portion of the insulin receptor. The beta subunit is composed of part of fibronectin domain III-2 and all of Fibronectin domain III-3. The beta subunit's FnIII-3 domain has links through the transmembrane region into the intracellular part of the membrane.Cryo-EM results have displayed clear representations of FnIII-2 and FnIII-3 domains, but lack in their ability to model the receptor structure throughout the transmembrane region and intracellular region. Although, the FnIII-3 domain is connected to these regions, so it has been proposed that the T-shape conformation extends all the way to the tyrosine kinase domain region. [https//www.rcsb.org/structure/1ir3 PDB 1IR3]]
Function
The insulin receptor's structure is critical to it's function. The receptor's ability to change shape is what initiates the signaling pathway that will eventually move glucose transporters to the cell surface, allowing glucose to passively defuse into the cell.
Activation by Insulin
Insulin is a peptide hormone produced and secreted from the islets of Langerhans of the pancreas in response to high blood glucose levels. Insulin is commonly considered the anabolic hormone of the body, and is the an important ligand in glucose homeostasis. The structure of is a simple monomer composed of two peptide chains linked by intermolecular disulfide bridges. Without insulin, the glucose receptors cannot be brought to the surface of the membrane to take in excess glucose in the blood, which means they are inactive. The body has a mechanism in place to upregulate the production of insulin in the presence of a surplus of glucose circulating in the blood stream. The binding of the increased amount of insulin to the insulin receptors will activate their downstream pathways to initiate glucose uptake by the phosphorylation of the Insulin Receptor Substrate (IRS). The transport of extracellular glucose into the cell allows it to be converted to glycogen for storage and later usage.
Conformation Change
When the receptor is in an shape, the FnIII-3 domains are separated by about 120Å. This distance prevents the initiation of autophosphorylation and downstream signaling by the tyrosine kinase domains on the intracellular side of the receptor. Upon the binding of insulin to three binding sites, 1, 1', and either 2 or 2', the conformation change will begin and bring the FnIII-3 domains within 40Å of each other to induce the conformation. [4] [3] The T shape conformation is well observed in the alpha subunit. It is horizontally composed of L1, CR (including the α-CT chain), and L2 domains and vertically composed of the FnIII-1, 2, and 3 domains. The insulin receptor's structural conformation change is what allows it to go from the inactive state to the active state in order to facilitate the autophosphorylation of the tyrosine kinase domain.
Binding interactions
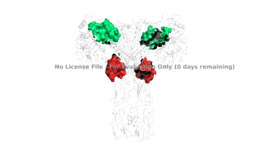
Figure 4: The presence of all four potential binding sides on the active insulin receptor: sites 1 and 1' (green) and sites 2, and 2'(red).
PDB 6SOFA tripartite interaction occurs between three critical parts of the alpha subunits of the insulin receptor. One one subunit, the α-CT chain and the FnIII-1 domain region become in close proximity during the conformational change of the insulin receptor. This interaction involves the following residues: and the . This duo then interacts with the leucine rich region, L1, that exists on the opposing alpha subunit of the dimer. The fact that the two alpha subunits are interacting displays a "cross linking" scenario where the domains of the heterodimer can intertwine with each other. The tripartite interaction between the α-CT chain, FnIII-1 domain, and the L1 region is important because it allows for a strong and stable interaction between two subunits of the insulin receptor that maintains the T-shape activation state for the rest of the downstream signaling to occur.
For insulin binding to induce the activation of the receptor and change its conformation to the active T state, binding at sites 1 and 1', as well as one insulin to either binding site 2 or 2', is required. [5]. Although interactions at all four binding sites are highly hydrophobic, the ligand binding interactions at sites 1 and 1' are different than at sites 2 and 2'. Sites 1 and 1' are signified by interactions between residues from the FnIII-1 domain and particular residues on the insulin ligand, such as HIS5. They also have significant disulfide linkages that help maintain a compact binging site. At sites 2 and 2' the FnIII-1 region has interacting with numerous residues on the surface of the insulin ligand.
Relevance
In a healthy individual, insulin is secreted in response to high blood glucose levels is bound by insulin receptors throughout the body which initiates a robust physiological response. Without properly functioning insulin receptors that can respond to increases in insulin, and therefore glucose, medical intervention is necessary for survival.
Disease
Diabetes mellitus is one of the most common diseases involving the insulin receptor in regards to glucose uptake and homeostasis. Type 1 diabetes is classified as "insulin dependent" and involves an inability for the body to produce insulin, resulting from damage or insufficiency in the Islets of Langerhans in the pancreas. Type 2 diabetes is classified as "insulin independent" and is the result of the body producing insufficient amounts of insulin, or not responding to the insulin. There are many molecular mechanisms that cause type 2 diabetes including lipotoxicity, hyperglycemia, and hyperinsulinemia which all cause hyper-activation of various protein phosphatases which reduces phosphorylation of the insulin receptor [6]. Type 1 diabetes is often treated with insulin injections, and type 2 diabetes is treated with diet and lifestyle changes [7] [8].
At the Cellular Level
The conformation change from the inactive to active state upon insulin binding has a time constant of six minutes. Autophosphorylation of the beta subunits reaches a maximal level in about one minute, and lasts for approximately six to ten minutes. One insulin receptor substrate has a half-life of 3.5 minutes where it is able to be phosphorylated by the tyrosine kinases of the beta subunit and then act as a central hub for further downstream signaling pathways. Once insulin binds to the alpha subunit, the receptor remains active for approximately ten minutes before the insulin is degraded and the receptor returns to its inactive conformation. This time frame puts a perspective on how long it takes for the human body to store excess glucose in their blood stream from a recent meal as glycogen for later use as fuel. [2]

