We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Harrison L. Smith/Sandbox1
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
====Alpha Subunits==== | ====Alpha Subunits==== | ||
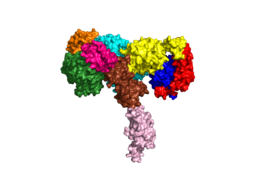
[[Image:Harrison Image2.png|thumb|right|260px|Figure 1: Insulin receptor apo receptor. Site L1' is colored a dark green, CR' is orange, L2' is bright blue, L2 is yellow, CR is red, L1 is dark blue, FnIII-1 is brown, and FnIII-2 is light pink. Insulin is shown bound and is colored dark pink. [http://www.rcsb.org/structure/6CE7 PDB 6CE7]]] | [[Image:Harrison Image2.png|thumb|right|260px|Figure 1: Insulin receptor apo receptor. Site L1' is colored a dark green, CR' is orange, L2' is bright blue, L2 is yellow, CR is red, L1 is dark blue, FnIII-1 is brown, and FnIII-2 is light pink. Insulin is shown bound and is colored dark pink. [http://www.rcsb.org/structure/6CE7 PDB 6CE7]]] | ||
| - | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. | + | The alpha subunits make up the extracellular domain ([http://en.wikipedia.org/wiki/Ectodomain ectodomain]) of the insulin receptor and are the sites of insulin binding. The alpha subunit is comprised of two Leucine rich domains (L1 & L2), a Cysteine rich domain (CR), and a C-terminal alpha helix. <ref name="Scapin"> PMID 29512653 </ref> The alpha subunits are held together by a [http://en.wikipedia.org/wiki/Disulfide disulfide bond] between <scene name='83/832953/Cysteine_bond/1'>cysteine residues</scene> at the CYS524 position on each alpha subunit. Two types of insulin binding sites are present in the alpha subunits, <scene name='83/832953/Sites_1_and_1_prime_location/15'>sites 1 and 1'</scene> and <scene name='83/832953/Sites_2_and_2_prime_location/12'>sites 2 and 2'</scene>. The sites are in pairs because of the heterodimeric nature of the receptor. Due to structural differences, as well as greater surface area and accessibility, binding sites 1 and 1' have much higher affinity than that of sites 2 and 2'. Insulin can also bind at sites 2 and 2', but the location on the back of the beta sheet of the FnIII-1 domain and lack of surface area decreases the likelihood of their binding site becoming occupied as quickly. <ref name="Uchikawa"> DOI 10.7554/eLife.48630 </ref> Cryo-EM has imaged insulin bound structures that displayed a T-shape conformation in the alpha subunits.<ref name="Uchikawa" /> |
[[Image:4 sites highlighted - Harrison.png|thumb|right|260px|Figure 2: The four binding sites of insulin. Sites 1 and 1' are colored green, sites 2 and 2' are colored red. [http://www.rcsb.org/structure/6SOF PDB 6SOF]]] | [[Image:4 sites highlighted - Harrison.png|thumb|right|260px|Figure 2: The four binding sites of insulin. Sites 1 and 1' are colored green, sites 2 and 2' are colored red. [http://www.rcsb.org/structure/6SOF PDB 6SOF]]] | ||
===Beta Subunits=== | ===Beta Subunits=== | ||
| - | The beta subunit spans from the extracellular domain across the transmembrane region and into the intracellular portion of the insulin receptor. The beta subunit is composed of part of [http://en.wikipedia.org/wiki/Fibronectin fibronectin] domain III-2 and all of Fibronectin domain III-3.<ref name="Scapin" | + | The beta subunit spans from the extracellular domain across the transmembrane region and into the intracellular portion of the insulin receptor. The beta subunit is composed of part of [http://en.wikipedia.org/wiki/Fibronectin fibronectin] domain III-2 and all of Fibronectin domain III-3. <ref name="Scapin" /> The beta subunit's FnIII-3 domain has links through the transmembrane region into the intracellular part of the membrane.[http://en.wikipedia.org/wiki/Transmission_electron_cryomicroscopy Cryo-EM] results have displayed clear representations of FnIII-2 and FnIII-3 domains, but lack in their ability to model the receptor structure throughout the transmembrane region and intracellular region. Although, the FnIII-3 domain is connected to these regions, so it has been proposed that the T-shape conformation extends all the way to the tyrosine kinase domain region. [http://www.rcsb.org/structure/1IR3 PDB 1IR3] |
== Function== | == Function== | ||
| Line 27: | Line 27: | ||
===Binding interactions=== | ===Binding interactions=== | ||
[[Image:4 sites highlighted.png|thumb|right|260px|Figure 4: The presence of all four potential binding sides on the active insulin receptor: sites 1 and 1' (green) and sites 2, and 2'(red). [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | [[Image:4 sites highlighted.png|thumb|right|260px|Figure 4: The presence of all four potential binding sides on the active insulin receptor: sites 1 and 1' (green) and sites 2, and 2'(red). [http://www.rcsb.org/structure/6sof PDB 6SOF]]] | ||
| - | A tripartite interaction occurs between three critical parts of the alpha subunits of the insulin receptor. One one subunit, the α-CT chain and the FnIII-1 domain region become in close proximity during the conformational change of the insulin receptor. This interaction involves the following residues: <scene name='83/832953/Alpha_ct_and_fniii-1/7'>ASP496, ARG498, and ASP499 on the FnIII-1 domain</scene> and the <scene name='83/832953/Alpha_ct_and_fniii-1/9'>LYS703, GLU706, and ASP707 on the α-CT domain</scene>. This duo then interacts with the leucine rich region, L1, that exists on the opposing alpha subunit of the dimer. The fact that the two alpha subunits are interacting displays a "cross linking" scenario where the domains of the heterodimer can intertwine with each other. The tripartite interaction between the α-CT chain, FnIII-1 domain, and the L1 region is important because it allows for a strong and stable interaction between two subunits of the insulin receptor that maintains the T-shape activation state for the rest of the downstream signaling to occur. | + | A <scene name='83/832953/Tripartite_interaction/1'>tripartite interaction</scene> occurs between three critical parts of the alpha subunits of the insulin receptor. <ref name="Uchikawa" /> One one subunit, the α-CT chain and the FnIII-1 domain region become in close proximity during the conformational change of the insulin receptor. This interaction involves the following residues: <scene name='83/832953/Alpha_ct_and_fniii-1/7'>ASP496, ARG498, and ASP499 on the FnIII-1 domain</scene> and the <scene name='83/832953/Alpha_ct_and_fniii-1/9'>LYS703, GLU706, and ASP707 on the α-CT domain</scene>. This duo then interacts with the leucine rich region, L1, that exists on the opposing alpha subunit of the dimer. The fact that the two alpha subunits are interacting displays a "cross linking" scenario where the domains of the heterodimer can intertwine with each other. The tripartite interaction between the α-CT chain, FnIII-1 domain, and the L1 region is important because it allows for a strong and stable interaction between two subunits of the insulin receptor that maintains the T-shape activation state for the rest of the downstream signaling to occur. <ref name="Uchikawa" /> |
For insulin binding to induce the activation of the receptor and change its conformation to the active T state, binding at sites 1 and 1', as well as one insulin to either binding site 2 or 2', is required. <ref> DOI 10.7554/eLife.48630 </ref>. Although interactions at all four binding sites are highly hydrophobic, the ligand binding interactions at sites 1 and 1' are different than at sites 2 and 2'. Sites 1 and 1' are signified by interactions between <scene name='83/832953/Sites_1_and_1_prime_location/14'>PRO495, PHE497, ARG498</scene> residues from the FnIII-1 domain and particular residues on the insulin ligand, such as HIS5. They also have significant disulfide linkages that help maintain a compact binging site. At sites 2 and 2' the FnIII-1 region has <scene name='83/832953/Sites_2_and_2_prime_location/10'>both basic residues-ARG479, LYS484, ARG488, ARG554- and hydrophobic residues- LEU486, LEU552, and PRO537-</scene> interacting with numerous residues on the surface of the insulin ligand. | For insulin binding to induce the activation of the receptor and change its conformation to the active T state, binding at sites 1 and 1', as well as one insulin to either binding site 2 or 2', is required. <ref> DOI 10.7554/eLife.48630 </ref>. Although interactions at all four binding sites are highly hydrophobic, the ligand binding interactions at sites 1 and 1' are different than at sites 2 and 2'. Sites 1 and 1' are signified by interactions between <scene name='83/832953/Sites_1_and_1_prime_location/14'>PRO495, PHE497, ARG498</scene> residues from the FnIII-1 domain and particular residues on the insulin ligand, such as HIS5. They also have significant disulfide linkages that help maintain a compact binging site. At sites 2 and 2' the FnIII-1 region has <scene name='83/832953/Sites_2_and_2_prime_location/10'>both basic residues-ARG479, LYS484, ARG488, ARG554- and hydrophobic residues- LEU486, LEU552, and PRO537-</scene> interacting with numerous residues on the surface of the insulin ligand. | ||
Revision as of 17:15, 19 April 2020
Homo sapiens Insulin Receptor
| |||||||||||
References
- ↑ 1.0 1.1 De Meyts P. The Insulin Receptor and Its Signal Transduction Network PMID:27512793
- ↑ 2.0 2.1 2.2 Tatulian SA. Structural Dynamics of Insulin Receptor and Transmembrane Signaling. Biochemistry. 2015 Sep 15;54(36):5523-32. doi: 10.1021/acs.biochem.5b00805. Epub , 2015 Sep 3. PMID:26322622 doi:http://dx.doi.org/10.1021/acs.biochem.5b00805
- ↑ 3.0 3.1 Scapin G, Dandey VP, Zhang Z, Prosise W, Hruza A, Kelly T, Mayhood T, Strickland C, Potter CS, Carragher B. Structure of the Insulin Receptor-Insulin Complex by Single Particle CryoEM analysis. Nature. 2018 Feb 28. pii: nature26153. doi: 10.1038/nature26153. PMID:29512653 doi:http://dx.doi.org/10.1038/nature26153
- ↑ 4.0 4.1 4.2 4.3 4.4 Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Weis F, Menting JG, Margetts MB, Chan SJ, Xu Y, Tennagels N, Wohlfart P, Langer T, Muller CW, Dreyer MK, Lawrence MC. The signalling conformation of the insulin receptor ectodomain. Nat Commun. 2018 Oct 24;9(1):4420. doi: 10.1038/s41467-018-06826-6. PMID:30356040 doi:http://dx.doi.org/10.1038/s41467-018-06826-6
- ↑ Uchikawa E, Choi E, Shang G, Yu H, Bai XC. Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex. Elife. 2019 Aug 22;8. pii: 48630. doi: 10.7554/eLife.48630. PMID:31436533 doi:http://dx.doi.org/10.7554/eLife.48630
- ↑ Boucher J, Kleinridders A, Kahn CR. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harb Perspect Biol. 2014 Jan 1;6(1). pii: 6/1/a009191. doi:, 10.1101/cshperspect.a009191. PMID:24384568 doi:http://dx.doi.org/10.1101/cshperspect.a009191
- ↑ Wilcox G. Insulin and insulin resistance. Clin Biochem Rev. 2005 May;26(2):19-39. PMID:16278749
- ↑ Riddle MC. Treatment of diabetes with insulin. From art to science. West J Med. 1983 Jun;138(6):838-46. PMID:6351440
Student Contributors
- Harrison Smith
- Alyssa Ritter