We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1619
From Proteopedia
(Difference between revisions)
| Line 20: | Line 20: | ||
===Substrate Structure=== | ===Substrate Structure=== | ||
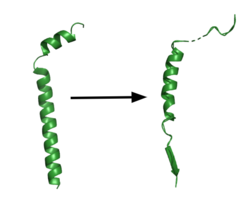
[[Image:App.png|250 px|right|thumb|'''Figure 1. APP fragment conformational change in gamma secretase.''' APP bound to gamma secretase undergoes a conformational change. The free state consists of 2 helices. The N-terminal helix unfolds into a coil and the C-terminal helix unwinds into a β-strand. This β-strand interacts with PS1 and is the site of cleavage by gamma secretase.]] | [[Image:App.png|250 px|right|thumb|'''Figure 1. APP fragment conformational change in gamma secretase.''' APP bound to gamma secretase undergoes a conformational change. The free state consists of 2 helices. The N-terminal helix unfolds into a coil and the C-terminal helix unwinds into a β-strand. This β-strand interacts with PS1 and is the site of cleavage by gamma secretase.]] | ||
| - | GS has multiple substrates, but the substrate of main concern is [https://en.wikipedia.org/wiki/Amyloid_precursor_protein/ APP]. APP is composed of an N-terminal loop and a transmembrane helix. It uses <scene name='83/832945/App_in_gs_general/1'>lateral diffusion</scene> as a mechanism of entry into the enzyme via the lid complex, and once in place, the TM helix is anchored by <scene name='83/832945/Hydrophobic_interactions/1'>van der Waals contacts in PS1</scene>. Upon binding to GS, the C-terminal extracellular helix unwinds. The N-terminal end of the TM helix unwinds into a β-strand (Fig. 1). In order to differentiate between different substrates like APP and notch, the β-strand is often the main point of identification for the enzyme. Substrate binding induces a structural change in GS, creating 2 β-strands that form a β-sheet with the substrate. This β-sheet is in close proximity with the active site, and guides the process of catalysis.<ref name="Zhou">PMID:30630874</ref> | + | GS has multiple substrates, but the substrate of main concern is [https://en.wikipedia.org/wiki/Amyloid_precursor_protein/ APP]. APP is composed of an N-terminal loop and a transmembrane helix. It uses <scene name='83/832945/App_in_gs_general/1'>lateral diffusion</scene> as a mechanism of entry into the enzyme via the lid complex, and once in place, the TM helix is anchored by <scene name='83/832945/Hydrophobic_interactions/1'>van der Waals contacts in PS1</scene>. Upon binding to GS, the C-terminal extracellular helix unwinds. The N-terminal end of the TM helix unwinds into a β-strand (Fig. 1). In order to differentiate between different substrates like APP and notch, the β-strand is often the main point of identification for the enzyme. Substrate binding induces a structural change in GS, creating 2 β-strands that form a β-sheet with the substrate. This β-sheet is in close proximity with the active site, and guides the process of catalysis.<ref name="Zhou">PMID:30630874</ref> |
Revision as of 00:23, 21 April 2020
Gamma Secretase
| |||||||||||
References
- ↑ 1.0 1.1 Yang G, Zhou R, Shi Y. Cryo-EM structures of human gamma-secretase. Curr Opin Struct Biol. 2017 Oct;46:55-64. doi: 10.1016/j.sbi.2017.05.013. Epub, 2017 Jul 17. PMID:28628788 doi:http://dx.doi.org/10.1016/j.sbi.2017.05.013
- ↑ 2.0 2.1 2.2 2.3 Zhou R, Yang G, Guo X, Zhou Q, Lei J, Shi Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science. 2019 Feb 15;363(6428). pii: science.aaw0930. doi:, 10.1126/science.aaw0930. Epub 2019 Jan 10. PMID:30630874 doi:http://dx.doi.org/10.1126/science.aaw0930
- ↑ 3.0 3.1 Bai XC, Yan C, Yang G, Lu P, Ma D, Sun L, Zhou R, Scheres SH, Shi Y. An atomic structure of human gamma-secretase. Nature. 2015 Aug 17. doi: 10.1038/nature14892. PMID:26280335 doi:http://dx.doi.org/10.1038/nature14892
- ↑ Bolduc DM, Montagna DR, Seghers MC, Wolfe MS, Selkoe DJ. The amyloid-beta forming tripeptide cleavage mechanism of gamma-secretase. Elife. 2016 Aug 31;5. doi: 10.7554/eLife.17578. PMID:27580372 doi:http://dx.doi.org/10.7554/eLife.17578
- ↑ Kumar D, Ganeshpurkar A, Kumar D, Modi G, Gupta SK, Singh SK. Secretase inhibitors for the treatment of Alzheimer's disease: Long road ahead. Eur J Med Chem. 2018 Mar 25;148:436-452. doi: 10.1016/j.ejmech.2018.02.035. Epub , 2018 Feb 15. PMID:29477076 doi:http://dx.doi.org/10.1016/j.ejmech.2018.02.035
Student Contributors
Layla Wisser
Daniel Mulawa