We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox DNA Polymerase Theta
From Proteopedia
(Difference between revisions)
(New page: <StructureSection load='4X0P' size='350' side='right' caption='DNA polymerase theta polymerase domain in complex with double-stranded DNA substrate and incoming ddATP (4X0P)' scene=''> ==...) |
|||
| Line 2: | Line 2: | ||
==Structural Description== | ==Structural Description== | ||
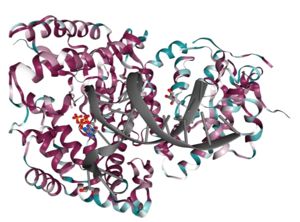
| - | The polymerase domain of DNA polymerase theta (<scene name='84/842971/Qm1/1'>QM1</scene>) is a member of the A-family of | + | The polymerase domain of [[DNA]] polymerase theta (<scene name='84/842971/Qm1/1'>QM1</scene>) is a member of the A-family of [[DNA polymerase]]s which includes [[DNA Polymerase I]] <ref>PMID: 8469987</ref> and T7 DNA polymerase <ref>PMID: 9440688</ref>. A-family polymerases contain <scene name='84/842971/A-family_motifs/1'>6 conserved motifs</scene>. These motifs allow for identification of A-family polymerases based on their amino acid sequence <ref>PMID: 8816490</ref>. In addition to the A-family motifs, the <scene name='84/842971/Finger-thumb-palm/1'>finger-palm-thumb</scene> subdomains that are typical of many polymerases are also conserved in QM1. The <scene name='84/842971/Fingers/1'>fingers subdomain</scene> coordinate the <scene name='84/842971/Dna/1'>DNA</scene> template and the <scene name='84/842971/Dds/1'>incoming nucleotide</scene> while the <scene name='84/842971/Palm/1'>palm subdomain</scene> performs catalysis and the <scene name='84/842971/Thumb/1'>thumb subdomain</scene> contacts the double stranded DNA. Within these subdomains exist disordered structural inserts that are not present in bacterial homologs. These inserts impart unique properties to QM1;<scene name='84/842971/Insert1/1'> insert 1</scene> in the thumb increases processivity while inserts <scene name='84/842971/Insert2/1'>2</scene> and <scene name='84/842971/Insert3/1'>3</scene> facilitate the ability of QM1 to bypass <scene name='84/842971/Thf/1'>DNA lesions</scene> that typically block synthesis by other A-family polymerases <ref>PMID: 21050863</ref>. This error prone synthesis<ref>PMID: 22135286</ref> makes QM1 more functionally similar to Y-family translesion polymerases. |
Beyond the canonical polymerase folds, QM1 also possesses a vestigial <scene name='84/842971/Exonuclease/1'>exonuclease</scene> domain that no longer performs proof-reading activities. This domain also contains <scene name='84/842971/Exo_1_2/1'>two structural inserts</scene> that are thought to contact the more distant helicase-like and central domains of full-length polymerase theta (see Function). | Beyond the canonical polymerase folds, QM1 also possesses a vestigial <scene name='84/842971/Exonuclease/1'>exonuclease</scene> domain that no longer performs proof-reading activities. This domain also contains <scene name='84/842971/Exo_1_2/1'>two structural inserts</scene> that are thought to contact the more distant helicase-like and central domains of full-length polymerase theta (see Function). | ||
| Line 21: | Line 21: | ||
==Relevance== | ==Relevance== | ||
| - | Polymerase theta over-expression in a number of cancers including breast cancer, non-small cell lung cancer, and oral squamous cell carcinoma is correlated with poor survival rates. Due to the role polymerase theta plays in double strand break repair, cells that over express polymerase theta are better able to repair damage caused by ionizing radiation treatment and chemotherapy. By developing compounds that inhibit polymerase theta activity in cancer cells, traditional cancer therapies can be made more effective in cancers that express high levels of polymerase theta. | + | Polymerase theta over-expression in a number of cancers including breast cancer <ref>PMID: 20624954</ref>, non-small cell lung cancer <ref>PMID: 23552402</ref>, and oral squamous cell carcinoma <ref>PMID: 22987617</ref> is correlated with poor survival rates. Due to the role polymerase theta plays in double strand break repair, cells that over express polymerase theta are better able to repair damage caused by ionizing radiation treatment and chemotherapy. By developing compounds that inhibit polymerase theta activity in cancer cells, traditional cancer therapies can be made more effective in cancers that express high levels of polymerase theta <ref>PMID: 27264557</ref>. |
| + | ==Relevant Structures== | ||
| + | [https://www.rcsb.org/structure/4X0Q Ternary complex of human DNA polymerase theta C-terminal domain binding ddGTP opposite dCMP] | ||
| + | [https://www.rcsb.org/structure/5A9J Crystal structure of the Helicase domain of human DNA polymerase theta, apo-form] | ||
| + | [https://www.rcsb.org/structure/1KFD Crystal structures of the Klenow fragment of DNA polymerase I complexed with deoxynucleoside triphosphate and pyrophosphate] | ||
| + | [https://www.rcsb.org/structure/1SKR T7 DNA Polymerase Complexed To DNA Primer/Template and ddATP] | ||
== References == | == References == | ||
<references/> | <references/> | ||
</StructureSection> | </StructureSection> | ||
Current revision
| |||||||||||