We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Angiotensin-Converting Enzyme
From Proteopedia
(Difference between revisions)
m (I updated the informatior about ACE2 receptor for COVID19 entry into the cell complementing that already existing for SARS-CoV in this page) |
|||
| Line 67: | Line 67: | ||
[[Treatments:Hypertension]]. | [[Treatments:Hypertension]]. | ||
| - | ==ACE2 and SARS== | + | ==ACE2 and coronavirus (SARS-CoV and COVID19) entry into the cell== |
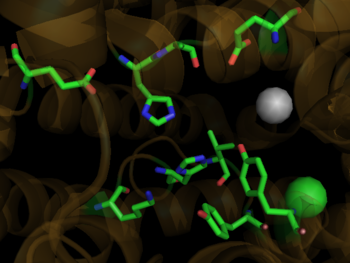
During the SARS scare of 2002-2003, extensive research was focused on the interactions between the SARS virus and its host cells. It was determined that the severe acute respiratory syndrome conavirus (SARS-CoV) enters cells through the activities of a spike shaped protein on its outer envelope. <ref name="SARS">PMID:18448527</ref> The Receptor Binding Domain (RBD) of SARS-CoV binds to ACE2, on the surface of the cell. It was determined that by changing a few selected residues on either the SARS-CoV RBD or the ACE2 binding site, the virus becomes significantly more infectious. <scene name='41/413151/Cv/1'>It is believed that these mutations</scene> ([[3d0g]]), namely at residues 31, 35, 38, & 353 in ACE2 or residues 479 and 487 in the SARS-CoV RBD, are what allowed for SARS transmission from [http://en.wikipedia.org/wiki/Civet Civets] to Humans. In fact, in those SARS strains which were determined to be most infectious, the unfavorable electrostatic interactions at the binding interface were removed via mutations at the critical residues 479 and 487. <ref name="SARS"/> | During the SARS scare of 2002-2003, extensive research was focused on the interactions between the SARS virus and its host cells. It was determined that the severe acute respiratory syndrome conavirus (SARS-CoV) enters cells through the activities of a spike shaped protein on its outer envelope. <ref name="SARS">PMID:18448527</ref> The Receptor Binding Domain (RBD) of SARS-CoV binds to ACE2, on the surface of the cell. It was determined that by changing a few selected residues on either the SARS-CoV RBD or the ACE2 binding site, the virus becomes significantly more infectious. <scene name='41/413151/Cv/1'>It is believed that these mutations</scene> ([[3d0g]]), namely at residues 31, 35, 38, & 353 in ACE2 or residues 479 and 487 in the SARS-CoV RBD, are what allowed for SARS transmission from [http://en.wikipedia.org/wiki/Civet Civets] to Humans. In fact, in those SARS strains which were determined to be most infectious, the unfavorable electrostatic interactions at the binding interface were removed via mutations at the critical residues 479 and 487. <ref name="SARS"/> | ||
Revision as of 10:16, 30 April 2020
| |||||||||||
Additional Resources
For Additional Information, see: Hypertension & Congestive Heart Failure
References
- ↑ Skeggs, L. T., Dorer, F. E., Kahn, J. R., Lentz, K. E., Levin, M. (1981) Experimental renal hypertension: the discovery of the Renin-Angiotensin system. Soffer, R. eds. Biochemical Regulation of Blood Pressure ,3-38 John Wiley & Sons, Inc. Hoboken.
- ↑ Hoogwerf BJ, Young JB. The HOPE study. Ramipril lowered cardiovascular risk, but vitamin E did not. Cleve Clin J Med. 2000 Apr;67(4):287-93. PMID:10780101
- ↑ 3.0 3.1 3.2 Ferrario CM. Role of angiotensin II in cardiovascular disease therapeutic implications of more than a century of research. J Renin Angiotensin Aldosterone Syst. 2006 Mar;7(1):3-14. PMID:17083068
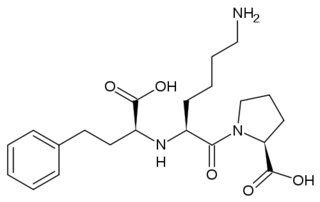
- ↑ Spyroulias GA, Nikolakopoulou P, Tzakos A, Gerothanassis IP, Magafa V, Manessi-Zoupa E, Cordopatis P. Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship. Eur J Biochem. 2003 May;270(10):2163-73. PMID:12752436
- ↑ 5.0 5.1 Brew K. Structure of human ACE gives new insights into inhibitor binding and design. Trends Pharmacol Sci. 2003 Aug;24(8):391-4. PMID:12915047
- ↑ 6.0 6.1 Sturrock ED, Natesh R, van Rooyen JM, Acharya KR. Structure of angiotensin I-converting enzyme. Cell Mol Life Sci. 2004 Nov;61(21):2677-86. PMID:15549168 doi:10.1007/s00018-004-4239-0
- ↑ 7.0 7.1 7.2 Weir MR. Effects of renin-angiotensin system inhibition on end-organ protection: can we do better? Clin Ther. 2007 Sep;29(9):1803-24. PMID:18035185 doi:10.1016/j.clinthera.2007.09.019
- ↑ Henriksen EJ, Jacob S. Modulation of metabolic control by angiotensin converting enzyme (ACE) inhibition. J Cell Physiol. 2003 Jul;196(1):171-9. PMID:12767053 doi:10.1002/jcp.10294
- ↑ Cole J, Ertoy D, Bernstein KE. Insights derived from ACE knockout mice. J Renin Angiotensin Aldosterone Syst. 2000 Jun;1(2):137-41. PMID:11967804
- ↑ Junot C, Gonzales MF, Ezan E, Cotton J, Vazeux G, Michaud A, Azizi M, Vassiliou S, Yiotakis A, Corvol P, Dive V. RXP 407, a selective inhibitor of the N-domain of angiotensin I-converting enzyme, blocks in vivo the degradation of hemoregulatory peptide acetyl-Ser-Asp-Lys-Pro with no effect on angiotensin I hydrolysis. J Pharmacol Exp Ther. 2001 May;297(2):606-11. PMID:11303049
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 Natesh R, Schwager SL, Sturrock ED, Acharya KR. Crystal structure of the human angiotensin-converting enzyme-lisinopril complex. Nature. 2003 Jan 30;421(6922):551-4. Epub 2003 Jan 19. PMID:12540854 doi:http://dx.doi.org/10.1038/nature01370
- ↑ Hangauer DG, Monzingo AF, Matthews BW. An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides. Biochemistry. 1984 Nov 20;23(24):5730-41. PMID:6525336
- ↑ Jaspard E, Alhenc-Gelas F. Catalytic properties of the two active sites of angiotensin I-converting enzyme on the cell surface. Biochem Biophys Res Commun. 1995 Jun 15;211(2):528-34. PMID:7794265
- ↑ http://www.yourlawyer.com/topics/overview/ace_inhibitors
- ↑ Natesh R, Schwager SL, Evans HR, Sturrock ED, Acharya KR. Structural details on the binding of antihypertensive drugs captopril and enalaprilat to human testicular angiotensin I-converting enzyme. Biochemistry. 2004 Jul 13;43(27):8718-24. PMID:15236580 doi:10.1021/bi049480n
- ↑ 16.0 16.1 Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol. 2008 Jul;82(14):6984-91. Epub 2008 Apr 30. PMID:18448527 doi:10.1128/JVI.00442-08
Proteopedia Page Contributors and Editors (what is this?)
David Canner, Michal Harel, Alexander Berchansky, Cristina Murga