Histone acetyltransferase 1-2 Complex (HAT1/2)
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
= HAT1/HAT2 Complex Structure = | = HAT1/HAT2 Complex Structure = | ||
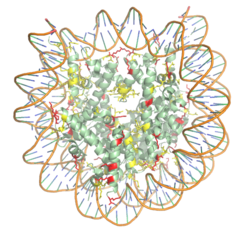
| - | The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> from the yeast ''Saccharomyces cerevisiae'' was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and a H4 N-terminal peptide (1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved. <ref name="Yang"/> | + | The structure of the <scene name='83/834210/Overview/2'>HAT1-HAT2 complex</scene> from the yeast ''Saccharomyces cerevisiae'' was determined by X-ray diffraction at 2.0Å resolution. The complex was crystallized in the presence of coenzyme A ([https://en.wikipedia.org/wiki/Coenzyme_A CoA]) and a H4 N-terminal peptide (1-48). The complex has four components (HAT1, HAT2, H4 peptide, and CoA) seen with a 1:1:1:1 stoichiometry. While residues 1-48 of the yeast H4 protein were included in the crystallization, only residues 9-46 were well resolved in the refined structure. Similarly, residues 1-4 in the HAT1 subunit and residues 1-6, 87-104, and 391-401 in the HAT2 subunit were not resolved.<ref name="Yang"/> |
| - | HAT1 is not catalytically active until it binds with HAT2 to form the complex. <ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl-coenzyme A. <scene name='83/834210/Hat2_-_chain_b/2'>HAT2</scene> is 401 residues in a beta-propeller formation with C7 symmetry. Bound to the HAT1-HAT2 complex is a <scene name='83/834210/Histone_4/3'>peptide substrate</scene> comprised of residues 9-46 of the histone protein H4. | + | HAT1 is not catalytically active until it binds with HAT2 to form the complex.<ref name="Wu"> PMID: 22615379 </ref> The <scene name='83/834210/Hat1_-_chain_a/2'>HAT1 structure</scene> includes 317 residues and contains the binding site for acetyl-coenzyme A. <scene name='83/834210/Hat2_-_chain_b/2'>HAT2</scene> is 401 residues in a beta-propeller formation with C7 symmetry. Bound to the HAT1-HAT2 complex is a <scene name='83/834210/Histone_4/3'>peptide substrate</scene> comprised of residues 9-46 of the histone protein H4. |
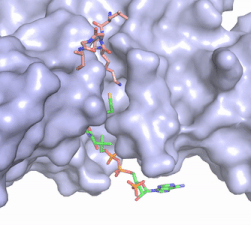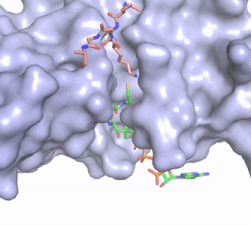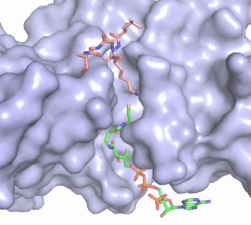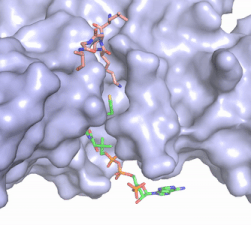
The HAT1 and HAT2 <scene name='81/811717/Salt_bridges/4'>interface</scene> is stabilized by multiple salt bridges, hydrogen bonds and hydrophobic van der Waals. Many of these are between a short helix spanning <scene name='83/834210/Lp1/3'>residues 200-208</scene> in HAT1 and several loops connecting β-strands in the propeller of HAT2. The HAT1 helix is important for the heterodimer formation as its deletion abolishes the ''in vitro'' interaction between HAT1 and HAT2.<ref name="Yang" /> Outside of the helix, three specific interactions at the interface involving hydrogen bonds help stabilize the complex: 1) The side chain atoms of <scene name='83/834210/Tyr199_asp308_ala202/4'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1, 2) The side chain of <scene name='83/834210/Lys211phe205_and_leu288arg282/3'>Lys211 and Arg282</scene> with main chain atoms of Leu288 and Phe205 respectively, and 3) Between the side-chains of <scene name='83/834210/Serine_hydrogen_bonds/2'>Ser263 and Asp 206</scene>. Finally, the <scene name='83/834210/Hydrophobic_core/2'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic residues from HAT1 and leucine residues from HAT2, however it does not form any obvious ring stacking. | The HAT1 and HAT2 <scene name='81/811717/Salt_bridges/4'>interface</scene> is stabilized by multiple salt bridges, hydrogen bonds and hydrophobic van der Waals. Many of these are between a short helix spanning <scene name='83/834210/Lp1/3'>residues 200-208</scene> in HAT1 and several loops connecting β-strands in the propeller of HAT2. The HAT1 helix is important for the heterodimer formation as its deletion abolishes the ''in vitro'' interaction between HAT1 and HAT2.<ref name="Yang" /> Outside of the helix, three specific interactions at the interface involving hydrogen bonds help stabilize the complex: 1) The side chain atoms of <scene name='83/834210/Tyr199_asp308_ala202/4'>Tyr199 and Asp308</scene> with the main chain nitrogen of Ala202 in HAT1, 2) The side chain of <scene name='83/834210/Lys211phe205_and_leu288arg282/3'>Lys211 and Arg282</scene> with main chain atoms of Leu288 and Phe205 respectively, and 3) Between the side-chains of <scene name='83/834210/Serine_hydrogen_bonds/2'>Ser263 and Asp 206</scene>. Finally, the <scene name='83/834210/Hydrophobic_core/2'>hydrophobic core</scene> at the interface of the complex appears to be critical for the complex formation. This core consists of aromatic residues from HAT1 and leucine residues from HAT2, however it does not form any obvious ring stacking. | ||
| Line 33: | Line 33: | ||
= Inhibition = | = Inhibition = | ||
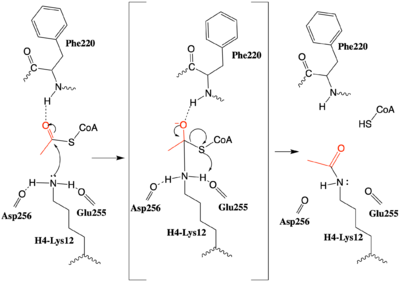
| - | Although HAT1 was the first histone acetyltransferase enzyme discovered, its function is poorly understood and the first inhibitor of the enzyme was only described in 2019<ref name=”Ngo”>PMID:30637990</ref>. The HAT1 inhibitor (H4K12CoA) that is a conjugate of the first 20 residues of the H4 protein, covalently linked to CoA through the lysine 12 side chain. H4K12CoA was found to act as a competitive inhibitor to both the H4 peptide substrate as well as acetyl-CoA. Having this inhibitor now allows for more in depth study of therapeutic targets associated with specific disease states. Additionally, H4K12CoA can be used as tool compound to better elucidate the HAT1 specificity and mechanism regarding epigenetic modification of the H4 histone protein. <ref name="Ngo"/> | + | Although HAT1 was the first histone acetyltransferase enzyme discovered, its function is poorly understood and the first inhibitor of the enzyme was only described in 2019<ref name=”Ngo”> PMID:30637990 </ref>. The HAT1 inhibitor (H4K12CoA) that is a conjugate of the first 20 residues of the H4 protein, covalently linked to CoA through the lysine 12 side chain. H4K12CoA was found to act as a competitive inhibitor to both the H4 peptide substrate as well as acetyl-CoA. Having this inhibitor now allows for more in depth study of therapeutic targets associated with specific disease states. Additionally, H4K12CoA can be used as tool compound to better elucidate the HAT1 specificity and mechanism regarding epigenetic modification of the H4 histone protein. <ref name="Ngo"/> |
= References = | = References = | ||
Revision as of 15:31, 28 May 2020
The Yeast HAT1-HAT2 Histone Acetyltransferase Complex Bound to a Histone H4 substrate
| |||||||||||
Student Contributors
Morgan Buckley, Jordan Finch, Caitlin Gaich, Kiran Kaur, Emily Leiderman, Ben Nick
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Angel Herraez, Michal Harel, Valentine J Klimkowski