WWP2
From Proteopedia
| Line 8: | Line 8: | ||
==Structure== | ==Structure== | ||
<StructureSection load='5TJ8' size='350' side='right' caption='WWP2 Ubiquitin Ligase Full-length Structure (PDB entry [[5TJ8]])' scene=''> | <StructureSection load='5TJ8' size='350' side='right' caption='WWP2 Ubiquitin Ligase Full-length Structure (PDB entry [[5TJ8]])' scene=''> | ||
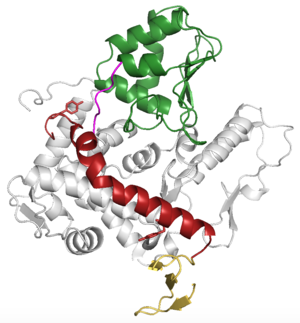
| - | WWP2 is a <scene name='84/848928/Overall/2'>monomer</scene> consisting of a C2 domain, four WW domains (labeled WW1-WW4), two lobes (labeled N and C), and a HECT domain. WWP2 has a 2,3-linker (shown in red) that is alpha helical in shape and connects WW2 and WW2 domains. This linker plays an autoinhibitory role. Upon phosphorylation the 2,3-linker changes conformation, allowing the protein to bind ubiquitin in the E2 binding site. This binding will further open the protein up to bind possible substrates. | + | WWP2 is a <scene name='84/848928/Overall/2'>monomer</scene> consisting of a C2 domain, four WW domains (labeled WW1-WW4), two lobes (labeled N and C), and a HECT domain. |
| + | |||
| + | WWP2 has a 2,3-linker (shown in red) that is alpha helical in shape and connects WW2 and WW2 domains. This linker plays an autoinhibitory role. Upon phosphorylation the 2,3-linker changes conformation, allowing the protein to bind ubiquitin in the E2 binding site. This binding will further open the protein up to bind possible substrates. | ||
| + | |||
The WW2 domain is a 3 stranded beta sheet. The HECT domain is in an inverse T shape when inactive and takes on an L shape when active. WW2 interaction with HECT is mediated by the six C terminal residues. | The WW2 domain is a 3 stranded beta sheet. The HECT domain is in an inverse T shape when inactive and takes on an L shape when active. WW2 interaction with HECT is mediated by the six C terminal residues. | ||
Revision as of 14:55, 16 June 2020
Contents |
Thoughts & Notes
Should we link this page to 5tjq (2,3-linker)? It also has a proteopedia page.
Introduction
WWP2 is a type of ubiquitin protein ligase. More specifically, it is a member of the Homologous to the E6-AP Carboxyl Terminus (HECT) E3 Ligases class which accept a ubiquitin molecule from an enzyme upstream in the ubiquitination pathway and transfer the ubiquitin to a Lysine residue in the target signaling molecule or transcription factor. The thioester bond formation between an active site Cystine on HECT E3 Ligases and ubiquitin differentiates this family of enzymes from the more abundant RING family of ubiquitin ligases which mediate ubiquitin transfer through non-covalent interactions. Within HECT E3 Ligases, WWP2 falls into the NEDD4 family. NEDD4 E3 Ligases consist of an N-terminal C2 Domain, between 2-4 WW domains, and a C-terminal HECT domain.
Structure
| |||||||||||
Function
Disease
Relevance
Structural highlights
References
1. A Tunable Brake for HECT Ubiquitin Ligases.,Chen Z, Jiang H, Xu W, Li X, Dempsey DR, Zhang X, Devreotes P, Wolberger C, Amzel LM, Gabelli SB, Cole PA Mol Cell. 2017 May 4;66(3):345-357.e6. doi: 10.1016/j.molcel.2017.03.020. PMID:2847587
Proteopedia Page Contributors and Editors (what is this?)
Tihitina Y Aytenfisu, Hannah Campbell, Sandra B. Gabelli, Michal Harel