User:Wally Novak/Sandbox2 RNAPolII
From Proteopedia
| Line 1: | Line 1: | ||
| - | |||
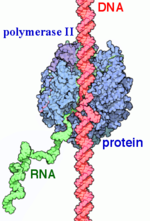
<StructureSection load='1i6h' size='340' side='right' caption='Yeast RNA Polymerase II complex with RNA (PDB code [[1i6h]])'> | <StructureSection load='1i6h' size='340' side='right' caption='Yeast RNA Polymerase II complex with RNA (PDB code [[1i6h]])'> | ||
== Introduction == | == Introduction == | ||
| Line 22: | Line 21: | ||
Transcription can largely be divided into three sections: initiation, elongation, and termination. In the process of initiation, RNAP II recruits several general transcription factors (GTFs) to bind to the promoter region of the DNA, and this eventually forms the preinitiation complex (PIC). The DNA enters RNAP II through the clamp, and then it is unwound, creating a transcription bubble. With the DNA unwound, the active site of RNAP II catalyzes the synthesis of the first few RNA bonds. Once the carboxy-terminal domain (CTD) becomes phosphorylated, the clamp undergoes a conformation change to effectively trap the DNA, and a few of the GTFs dissociate, which changes complex to the Elongator complex. | Transcription can largely be divided into three sections: initiation, elongation, and termination. In the process of initiation, RNAP II recruits several general transcription factors (GTFs) to bind to the promoter region of the DNA, and this eventually forms the preinitiation complex (PIC). The DNA enters RNAP II through the clamp, and then it is unwound, creating a transcription bubble. With the DNA unwound, the active site of RNAP II catalyzes the synthesis of the first few RNA bonds. Once the carboxy-terminal domain (CTD) becomes phosphorylated, the clamp undergoes a conformation change to effectively trap the DNA, and a few of the GTFs dissociate, which changes complex to the Elongator complex. | ||
| - | After initiation, the process of elongation begins with the entry of NTPs. These NTPs largely enter through the funnel, and this tends to be a slow process because the funnel is only 12 Å in diameter, meaning that only one NTP can go through at a time. Once they have passed through the funnel, the NTPs enter <scene name=' | + | After initiation, the process of elongation begins with the entry of NTPs. These NTPs largely enter through the funnel, and this tends to be a slow process because the funnel is only 12 Å in diameter, meaning that only one NTP can go through at a time. Once they have passed through the funnel, the NTPs enter <scene name='86/860989/Active_site_wn_test/1'>active site</scene> which contains an Mg ion that is bound by three aspartate residues at positions D739, D741, and D743. If the NTP is complementary to the DNA strand, it is loaded onto the insertion site next to the RNA and held in place by the bridge. While in this insertion site, the metal ion and aspartate residues catalyze the reaction that forms a phosphodiester bond between the 3’ end of the RNA and the 5’ end of the NTP. This step then leads into a translocation step. To make room for the next NTP, there is a Brownian ratchet mechanism in which the nearby trigger loop undergoes a conformation change that causes the bridge to move to the next transition site. During the bridge’s transition, the DNA-RNA hybrid is partially help in place by the -amanitin. Once the bridge is in the new initiation site, the trigger loop returns to its original conformation, allowing the process to begin again. |
In the termination stage, the CTD becomes dephosphorylated, which acts as a signal to dissociate the elongation complex. Once this signal is received, the new mRNA is released from RNP II through the rudder. Before being released, its 3’ end is polyadenylated. The DNA is brought back together at the other end of the transcription bubble, returning it to its original double-stranded form. Finally, RNAP II and the remaining GTFs dissociate from the DNA. | In the termination stage, the CTD becomes dephosphorylated, which acts as a signal to dissociate the elongation complex. Once this signal is received, the new mRNA is released from RNP II through the rudder. Before being released, its 3’ end is polyadenylated. The DNA is brought back together at the other end of the transcription bubble, returning it to its original double-stranded form. Finally, RNAP II and the remaining GTFs dissociate from the DNA. | ||
Current revision
| |||||||||||
References
Bushnell, D. A.; Westover, K. D.; Davis, R. E.; Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II-TFIIB Cocrystal at 4.5 Angstroms. Science. 2004, 303, 983-988
Brueckner, F. and Cramer, P. Structural Basis of Transcription Inhibition by -amanitin and Implications for RNA Polymerase II Translocation. Nature Structure and Molecular Biology. 2008, 15, 811-818.
Cramer, P.; Bushnell, D. A.; Kornberg, R. D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Ångstrom Resolution. Science. 2001, 292, 1863-1876
Evans, D. A.; Fitch, D. M.; Smith, T. E.; Cee, V. J. Application of Complex Aldol Reactions to the Total Synthesis of Phorboxazole B. J. Am. Chem. Soc. 2000, 122, 10033-10046.
Gnatt, A. L.; Cramer, P; Fu, J.; Bushnell, D. A.; and Kornberg, R. D. Structural Basis of Transcription: An RNA Polymerase II Elongation Complex at 3.3 Å Resolution. Science. 2001, 292, 1876-1882 1i6h
Hahn, S. Structure and Mechanism of the RNA Polymerase II Transcription Machinery. Nature Structure and Molecular Biology. 2004, 11, 394-403.
He, Yuan, et al. Near-atomic resolution visualization of human transcription promoter opening. Nature 533.7603. 2016.
Nudler, E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009, 78, 335-361.
Orphanides, George, Thierry Lagrange, and Danny Reinberg. The general transcription factors of RNA polymerase II. Genes & development 10.21. 1996. 2657-2683
Shah, N. et. al. Tyrosine-1 of RNA Polymerase II CTD Controls Global Termination of Gene Transcription in Mammals. Molecular Cell. 2018, 69, 48-61.
Uzman, A.; Voet, D. Student companion Fundamentals of biochemistry: life at the molecular level, 4th ed., Donald Voet, Judith G. Voet, Charlotte W. Pratt; John Wiley & amp; Sons, 2012.
Xu, J.; Lahiri, I.; Wang, W.; Wier, A.; Cianfrocco, M. A.; Chong, J.; Hare, A. A.; Dervan, P. B.; DiMaio, F.; Leschziner, A. E.; Wang, D. Structural Basis for the Initiation of Eukaryotic Transcription-coupled DNA Repair. Nature. 2017. 551, 653-657 5vvr
Xin, L.; Bushnell, D. A.; and Kornburg, R. D. RNA Polymerase II Transcription: Structure and Mechanism. Biochemica et Biophysica Acta. 2013, 1829, 2-8.
Yan, C., Dodd, T., He, Y., Tainer, J. A., Tsutakawa, S. E., & Ivanov, I. (2019). Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nature Structural and Molecular Biology, 26(6), 397-406.
Alpha-aminitin chemical structure image courtesy of https://en.wikipedia.org/wiki/Alpha-Amanitin#/media/File:Alpha-amanitin_structure.png
Notes
From structural components:
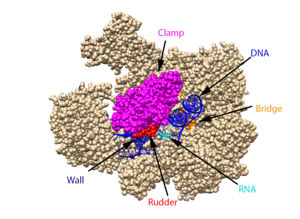
Structural overview: [PDB: 5VVR: with highlighted sections mentioned below]
Bridge: Depicted: [PDB: 1I6H: 810-845.a]
Wall: Depicted: [PDB: 1R5U: 853-919.b; 933-972.b]
Clamp: Depicted: [PDB: 1R5U: 3-345.a; 1395-1435.a; 1158-1124.b]
Rudder: Depicted: [PDB: 5VVR: 306-321.a]
Content Donators
This page was created as a final project for the Advanced Biochemistry course at Wabash College during the Fall of 2019. This page was reviewed by Dr. Wally Novak of Wabash College.