Hypoxia-Inducible Factors
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | < | + | <StructureSection load='4ZPK' size='350' side='right' caption='Insert caption here' scene='Insert optional scene name here' > |
| + | |||
'''Hypoxia-inducible factors''' ('''HIFs''') are transcription factors responsible of the cellular adaptation to [https://en.wikipedia.org/wiki/Hypoxia_(medical) hypoxia], which is a condition of low oxygen availability. Among the genes regulated by HIF, we can find those involved in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] and [https://en.wikipedia.org/wiki/Metabolism metabolism]. | '''Hypoxia-inducible factors''' ('''HIFs''') are transcription factors responsible of the cellular adaptation to [https://en.wikipedia.org/wiki/Hypoxia_(medical) hypoxia], which is a condition of low oxygen availability. Among the genes regulated by HIF, we can find those involved in [https://en.wikipedia.org/wiki/Erythropoiesis erythropoiesis], [https://en.wikipedia.org/wiki/Angiogenesis angiogenesis] and [https://en.wikipedia.org/wiki/Metabolism metabolism]. | ||
==Structure <ref>PMID: 16887934</ref><ref>PMID: 26245371</ref>== | ==Structure <ref>PMID: 16887934</ref><ref>PMID: 26245371</ref>== | ||
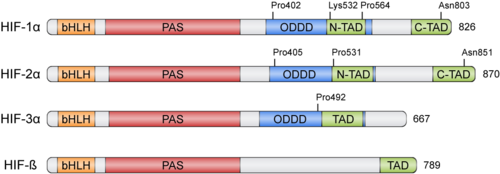
| - | HIFs were discovered more than 30 years ago thanks to the [[Erythropoietin]] (''EPO'') gene, an hormone that is transcribed in hypoxic conditions and stimulates erythrocyte proliferation <ref>PMID: 2849206</ref>. ''EPO'' presents an upstream '''Hypoxia Response Element''' ('''HRE''') that resulted to be bound by HIF <ref>PMID: 2062846</ref>. HIFs form part of the '''basic helix-loop-helix-Per-ARNT-Sim''' ([https://en.wikipedia.org/wiki/Basic_helix-loop-helix bHLH]-[https://en.wikipedia.org/wiki/PAS_domain PAS]) family of proteins<ref>PMID: 7539918</ref>. Active HIFs are achieved by heterodimerization of a constitutively expressed subunit '''HIF-β''' and an oxygen-regulated subunit, which can be '''HIF-1α''' or its paralogs HIF-2α and HIF-3α (Table 1.). HIF-β is also known as the '''aryl hydrocarbon nuclear translocator''' ('''ARNT''')<ref>PMID: 1317062</ref>. Their structure can contain the following domains from the N-terminus to the C-terminus: | + | HIFs were discovered more than 30 years ago thanks to the [[Erythropoietin]] (''EPO'') gene, an hormone that is transcribed in hypoxic conditions and stimulates erythrocyte proliferation<ref>PMID: 2849206</ref>. ''EPO'' presents an upstream '''Hypoxia Response Element''' ('''HRE''') that resulted to be bound by HIF <ref>PMID: 2062846</ref>. HIFs form part of the '''basic helix-loop-helix-Per-ARNT-Sim''' ([https://en.wikipedia.org/wiki/Basic_helix-loop-helix bHLH]-[https://en.wikipedia.org/wiki/PAS_domain PAS]) family of proteins<ref>PMID: 7539918</ref>. Active HIFs are achieved by heterodimerization of a constitutively expressed subunit '''HIF-β''' and an oxygen-regulated subunit, which can be '''HIF-1α''' or its paralogs HIF-2α and HIF-3α (Table 1.). HIF-β is also known as the '''aryl hydrocarbon nuclear translocator''' ('''ARNT''')<ref>PMID: 1317062</ref>. Their structure can contain the following domains from the N-terminus to the C-terminus: |
* basic Helix-Loop-Helix ('''bHLH'''): essential for heterodimer formation and DNA binding to HRE | * basic Helix-Loop-Helix ('''bHLH'''): essential for heterodimer formation and DNA binding to HRE | ||
| Line 10: | Line 11: | ||
* N-terminal and C-terminal transactivating domains ('''N-TAD''' and '''C-TAD'''): bind different coactivators to promote gene expression, such as [https://en.wikipedia.org/wiki/P300-CBP_coactivator_family p300/CBP]<ref>PMID: 11959977</ref>. HIF-3α lacks one of the TAD domains. | * N-terminal and C-terminal transactivating domains ('''N-TAD''' and '''C-TAD'''): bind different coactivators to promote gene expression, such as [https://en.wikipedia.org/wiki/P300-CBP_coactivator_family p300/CBP]<ref>PMID: 11959977</ref>. HIF-3α lacks one of the TAD domains. | ||
| + | [[Image: HIF members structure GonzaloGarcia-Martin 2020.png|500 px|thumb|left|HIF members structure]] | ||
{| class="wikitable" style="text-align: left;" | {| class="wikitable" style="text-align: left;" | ||
|- | |- | ||
| Line 31: | Line 33: | ||
|} | |} | ||
| - | [[File: HIF members structure GonzaloGarcia-Martin 2020.png | thumb | left | alt= HIF members structure]] | ||
==Physiological Function== | ==Physiological Function== | ||
| Line 56: | Line 57: | ||
</StructureSection> | </StructureSection> | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 18:36, 14 November 2020
| |||||||||||
References
- ↑ Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1). Mol Pharmacol. 2006 Nov;70(5):1469-80. doi: 10.1124/mol.106.027029. Epub 2006 Aug, 3. PMID:16887934 doi:http://dx.doi.org/10.1124/mol.106.027029
- ↑ Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015 Aug 5. doi: 10.1038/nature14883. PMID:26245371 doi:http://dx.doi.org/10.1038/nature14883
- ↑ Goldberg MA, Dunning SP, Bunn HF. Regulation of the erythropoietin gene: evidence that the oxygen sensor is a heme protein. Science. 1988 Dec 9;242(4884):1412-5. doi: 10.1126/science.2849206. PMID:2849206 doi:http://dx.doi.org/10.1126/science.2849206
- ↑ Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5680-4. doi: 10.1073/pnas.88.13.5680. PMID:2062846 doi:http://dx.doi.org/10.1073/pnas.88.13.5680
- ↑ Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995 Jun 6;92(12):5510-4. PMID:7539918
- ↑ Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992 May 22;256(5060):1193-5. PMID:1317062
- ↑ Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002 Apr 16;99(8):5271-6. PMID:11959977 doi:http://dx.doi.org/10.1073/pnas.082121399
- ↑ Loboda A, Jozkowicz A, Dulak J. HIF-1 and HIF-2 transcription factors--similar but not identical. Mol Cells. 2010 May;29(5):435-42. doi: 10.1007/s10059-010-0067-2. Epub 2010 Apr, 12. PMID:20396958 doi:http://dx.doi.org/10.1007/s10059-010-0067-2
- ↑ Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005 Jan 15;105(2):659-69. doi: 10.1182/blood-2004-07-2958. Epub 2004 Sep , 16. PMID:15374877 doi:http://dx.doi.org/10.1182/blood-2004-07-2958
- ↑ Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001 Nov 9;294(5545):1337-40. doi: 10.1126/science.1066373. Epub 2001, Oct 11. PMID:11598268 doi:http://dx.doi.org/10.1126/science.1066373
- ↑ Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci U S A. 2000 Sep 12;97(19):10430-5. PMID:10973499 doi:http://dx.doi.org/10.1073/pnas.190332597
- ↑ Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1alpha by ARD1-mediated acetylation. Cell. 2002 Nov 27;111(5):709-20. PMID:12464182
- ↑ Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002 Feb 1;295(5556):858-61. doi: 10.1126/science.1068592. PMID:11823643 doi:http://dx.doi.org/10.1126/science.1068592
- ↑ Richard DE, Berra E, Gothie E, Roux D, Pouyssegur J. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1alpha (HIF-1alpha) and enhance the transcriptional activity of HIF-1. J Biol Chem. 1999 Nov 12;274(46):32631-7. doi: 10.1074/jbc.274.46.32631. PMID:10551817 doi:http://dx.doi.org/10.1074/jbc.274.46.32631
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Proteopedia Page Contributors and Editors (what is this?)
Gonzalo Garcia-Martin, Michal Harel, Carmen Paredes Yubero, Rocío Moreno