We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
==The Spike Protein== | ==The Spike Protein== | ||
The S-protein is a structural protein extends from the viral membrane and it is uniformly arranged as a trimer on the surface to give the crown-like appearance of the SARS-Co | The S-protein is a structural protein extends from the viral membrane and it is uniformly arranged as a trimer on the surface to give the crown-like appearance of the SARS-Co | ||
| - | V-2. The Spike protein is 14-1255bp long, which mediates a receptor binding and fusion of the virus and a cellular membrane <ref>Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033</ref>. Is also gives the virus its name in Latin as "Corona". | + | V-2. The Spike protein is 14-1255bp long, which mediates a receptor binding Val367<scene name='75/752266/Val367/1'>Binding region val367</scene> and fusion of the virus and a cellular membrane <ref>Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033</ref>. Is also gives the virus its name in Latin as "Corona". |
==Domains== | ==Domains== | ||
| - | The S-protein of the SARS-CoV-2 has two main subunits , an S1 and S2 subunits. The S1 subunit is located on residue #14–685 ( contains the NTD) and interact with human AEC2 by attaching its virion to the cell membrane by interacting with host receptor. The S2 subunit is located on residue #686–1273 ( contains the CT) which serves as the fusion protein of the virus <ref>UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594</ref>. | + | The S-protein of the SARS-CoV-2 has two main subunits , an S1 and S2 subunits NTD-CT<scene name='75/752266/Ntd_-_ct/1'>S1 subunit is in the downstream of NTD and S2 subunit is in the upsteam of CT</scene> . The S1 subunit is located on residue #14–685 ( contains the NTD) and interact with human AEC2 by attaching its virion to the cell membrane by interacting with host receptor. The S2 subunit is located on residue #686–1273 ( contains the CT) which serves as the fusion protein of the virus <ref>UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594</ref>. |
==Activation of S-protein== | ==Activation of S-protein== | ||
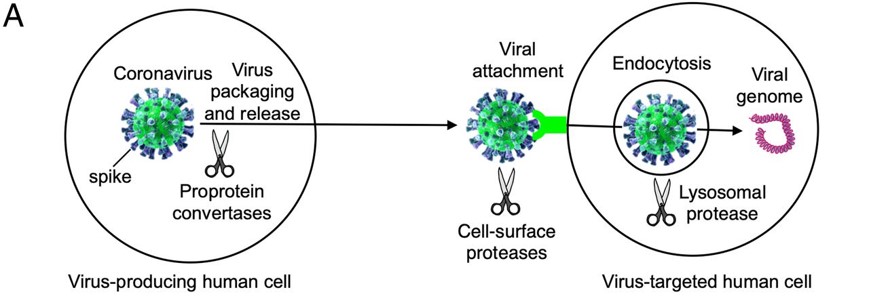
| - | Before the Spike protein can be activated , it has to be cleaved by the protease Furin protein. This 2D image below shows the schematic cleavage of the S-protein before and after it enters the host cell | + | Before the Spike protein can be activated , it has to be cleaved by the protease Furin protein ALA 668 <scene name='75/752266/Ala_668/1'>Furin Cleavage site</scene> . This 2D image below shows the schematic cleavage of the S-protein before and after it enters the host cell |
[[Image:Cleavage.jpg]] <ref>Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020, May 26). Cell entry mechanisms of SARS-CoV-2. Retrieved November 14, 2020, from https://www.pnas.org/content/117/21/11727</ref> | [[Image:Cleavage.jpg]] <ref>Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020, May 26). Cell entry mechanisms of SARS-CoV-2. Retrieved November 14, 2020, from https://www.pnas.org/content/117/21/11727</ref> | ||
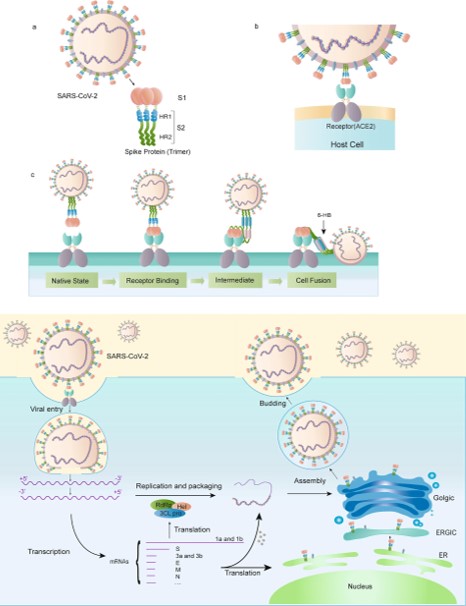
==The Mechanism of the Spike Protein== | ==The Mechanism of the Spike Protein== | ||
| + | LYS 187 <scene name='75/752266/Lys_187/1'>The Receptor Binding Domain (RBD)</scene> | ||
[[Image:Picture3.jpg]]<ref>Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4</ref> | [[Image:Picture3.jpg]]<ref>Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4</ref> | ||
| Line 26: | Line 27: | ||
==Mutation== | ==Mutation== | ||
| - | The most studied mutation site of the S-protein is at residue 614 which encodes for the amino acid Aspartic acid (D) and is normally changed to Glycine (G). And this form of mutation causes the enhancement of the viral transmission <ref>Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033</ref>. | + | The most studied mutation site of the S-protein is at residue 614 which encodes for the amino acid Aspartic acid (D) D614 <scene name='75/752266/Asp_614/1'>The Mutation site D614 </scene> and is normally changed to Glycine (G). And this form of mutation causes the enhancement of the viral transmission <ref>Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033</ref>. |
| Line 33: | Line 34: | ||
== Structural highlights == | == Structural highlights == | ||
| - | These are the structural 3D representations of the s-protein showing the two subunits , the binding regions to its receptor human ACE2 , Mutation site , RBD site and cleavage site respectively NTD -CT<scene name='75/752266/Ntd_-_ct/1'>S1 subunit is in the downstream of NTD and S2 subunit is in the upsteam of CT</scene> .Val367<scene name='75/752266/Val367/1'>Binding region val367</scene> . D614 <scene name='75/752266/Asp_614/1'>The Mutation site D614 </scene>. LYS 187 <scene name='75/752266/Lys_187/1'>The Receptor Binding Domain (RBD)</scene>.ALA 668 <scene name='75/752266/Ala_668/1'>Furin Cleavage site</scene> | + | These are the structural 3D representations of the s-protein showing the two subunits , the binding regions to its receptor human ACE2 , Mutation site , RBD site and cleavage site respectively NTD-CT<scene name='75/752266/Ntd_-_ct/1'>S1 subunit is in the downstream of NTD and S2 subunit is in the upsteam of CT</scene> .Val367<scene name='75/752266/Val367/1'>Binding region val367</scene> . D614 <scene name='75/752266/Asp_614/1'>The Mutation site D614 </scene>. LYS 187 <scene name='75/752266/Lys_187/1'>The Receptor Binding Domain (RBD)</scene>.ALA 668 <scene name='75/752266/Ala_668/1'>Furin Cleavage site</scene> |
</StructureSection> | </StructureSection> | ||
Revision as of 13:51, 16 November 2020
Spike glycoprotein
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4
- ↑ Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033
- ↑ UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. (2020, October 07). Spike glycoprotein. Retrieved November 13, 2020, from https://www.uniprot.org/uniprot/P59594
- ↑ Shang, J., Wan, Y., Luo, C., Ye, G., Geng, Q., Auerbach, A., & Li, F. (2020, May 26). Cell entry mechanisms of SARS-CoV-2. Retrieved November 14, 2020, from https://www.pnas.org/content/117/21/11727
- ↑ Huang, Y., Yang, C., Xu, X., Xu, W., & Liu, S. (2020). Structural and functional properties of SARS-CoV-2 spike protein: Potential antivirus drug development for COVID-19. Acta Pharmacologica Sinica, 41(9), 1141-1149. doi:10.1038/s41401-020-0485-4
- ↑ Mohammad, A., Alshawaf, E., Marafie, S. K., Abu-Farha, M., Abubaker, J., & Al-Mulla, F. (2020). Higher binding affinity of Furin to SARS-CoV-2 spike (S) protein D614G could be associated with higher SARS-CoV-2 infectivity. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases, S1201-9712(20)32237-2. Advance online publication. https://doi.org/10.1016/j.ijid.2020.10.033