1tf7
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
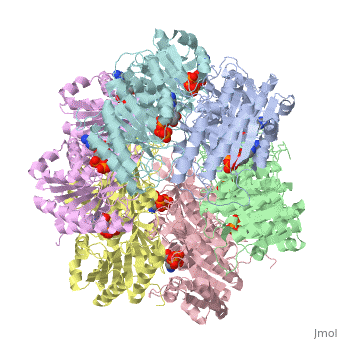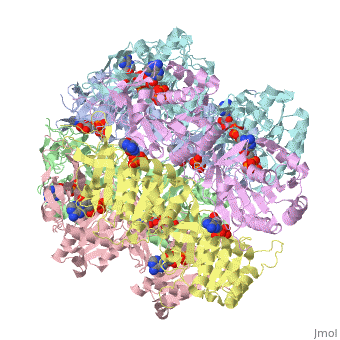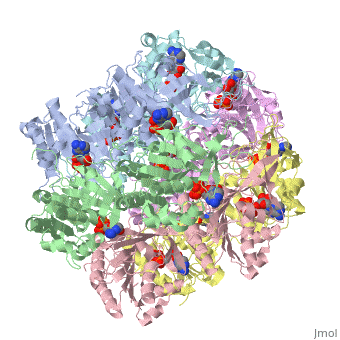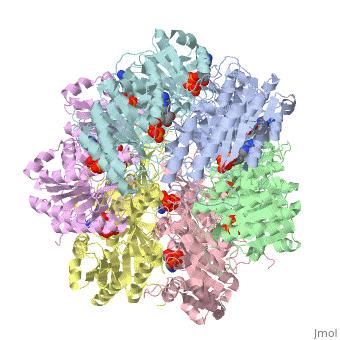
<StructureSection load='1tf7' size='340' side='right'caption='[[1tf7]], [[Resolution|resolution]] 2.80Å' scene=''> | <StructureSection load='1tf7' size='340' side='right'caption='[[1tf7]], [[Resolution|resolution]] 2.80Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1tf7]] is a 6 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1tf7]] is a 6 chain structure with sequence from [https://en.wikipedia.org/wiki/Synechococcus_sp. Synechococcus sp.]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1TF7 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1TF7 FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.8Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ATP:ADENOSINE-5-TRIPHOSPHATE'>ATP</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1tf7 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1tf7 OCA], [https://pdbe.org/1tf7 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1tf7 RCSB], [https://www.ebi.ac.uk/pdbsum/1tf7 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1tf7 ProSAT]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/KAIC_SYNE7 KAIC_SYNE7] Core component of the KaiABC clock protein complex, which constitutes the main circadian regulator in cyanobacteria. Binds to DNA. The KaiABC complex may act as a promoter-nonspecific transcription regulator that represses transcription, possibly by acting on the state of chromosome compaction.<ref>PMID:9727980</ref> <ref>PMID:14709675</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 20: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1tf7 ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1tf7 ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | Circadian (daily) biological clocks express characteristics that are difficult to explain by known biochemical mechanisms, and will ultimately require characterizing the structures, functions, and interactions of their molecular components. KaiC is an essential circadian protein in cyanobacteria that forms the core of the KaiABC clock protein complex. We report the crystal structure of the KaiC homohexameric complex at 2.8 A resolution. The structure resembles a double doughnut with a central pore that is partially sealed at one end. The crystal structure reveals ATP binding, inter-subunit organization, a scaffold for Kai-protein complex formation, the location of critical KaiC mutations, and evolutionary relationships to other proteins. A key auto-phosphorylation site on KaiC (T432) is identified from the crystal structure, and mutation of this residue abolishes circadian rhythmicity. The crystal structure of KaiC will be essential for understanding this circadian clockwork and for establishing its links to global gene expression. | ||
| - | |||
| - | Visualizing a circadian clock protein: crystal structure of KaiC and functional insights.,Pattanayek R, Wang J, Mori T, Xu Y, Johnson CH, Egli M Mol Cell. 2004 Aug 13;15(3):375-88. PMID:15304218<ref>PMID:15304218</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1tf7" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| - | *[[Circadian Clock Protein KaiC|Circadian Clock Protein KaiC]] | ||
*[[Circadian clock protein 3D structures|Circadian clock protein 3D structures]] | *[[Circadian clock protein 3D structures|Circadian clock protein 3D structures]] | ||
== References == | == References == | ||
| Line 38: | Line 28: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: | + | [[Category: Synechococcus sp]] |
| - | [[Category: Egli | + | [[Category: Egli M]] |
| - | [[Category: Johnson | + | [[Category: Johnson CH]] |
| - | [[Category: Mori | + | [[Category: Mori T]] |
| - | [[Category: Pattanayek | + | [[Category: Pattanayek R]] |
| - | [[Category: Wang | + | [[Category: Wang J]] |
| - | [[Category: Xu | + | [[Category: Xu Y]] |
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal Structure of Circadian Clock Protein KaiC
| |||||||||||
Categories: Large Structures | Synechococcus sp | Egli M | Johnson CH | Mori T | Pattanayek R | Wang J | Xu Y