This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
PROTAC
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | PROTAC ( | + | PROTAC (proteolysis-targeting chimera) is a small protein with two active domains joined by a linker. |
[[Image:PROTAC_70.png|thumb|right|300px|Representative PROTAC from [https://protacpedia.weizmann.ac.il/ptcb/detp?i=70 PROTACpedia] targeting EGFR Epidermal growth factor receptor P00533. <span class="bg-lightblue"><b>target binding ligand</b></span><span class="bg-lightmagenta"><b>linker</b></span><span class="bg-lightgreen"><b>E3 binder</b></span>]] | [[Image:PROTAC_70.png|thumb|right|300px|Representative PROTAC from [https://protacpedia.weizmann.ac.il/ptcb/detp?i=70 PROTACpedia] targeting EGFR Epidermal growth factor receptor P00533. <span class="bg-lightblue"><b>target binding ligand</b></span><span class="bg-lightmagenta"><b>linker</b></span><span class="bg-lightgreen"><b>E3 binder</b></span>]] | ||
| + | |||
| + | ==Predicting PROTACs== | ||
| + | PRosettaC <ref>PRosettaC https://prosettac.weizmann.ac.il</ref> is a computational protocol for the prediction of PROTAC-induced ternary complexes <ref>pmid 32976709</ref>. The protocol receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex. | ||
| + | |||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 07:57, 28 February 2021
PROTAC (proteolysis-targeting chimera) is a small protein with two active domains joined by a linker.
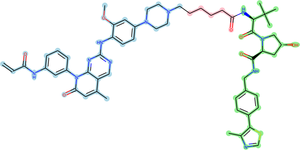
Representative PROTAC from PROTACpedia targeting EGFR Epidermal growth factor receptor P00533. target binding ligandlinkerE3 binder
Predicting PROTACs
PRosettaC [1] is a computational protocol for the prediction of PROTAC-induced ternary complexes [2]. The protocol receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex.
References
- ↑ PRosettaC https://prosettac.weizmann.ac.il
- ↑ Zaidman D, Prilusky J, London N. PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model. 2020 Sep 25. doi: 10.1021/acs.jcim.0c00589. PMID:32976709 doi:http://dx.doi.org/10.1021/acs.jcim.0c00589
