PROTAC
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
==Predicting PROTACs== | ==Predicting PROTACs== | ||
PRosettaC <ref>PRosettaC https://prosettac.weizmann.ac.il</ref> is a computational protocol for the prediction of PROTAC-induced ternary complexes <ref>pmid 32976709</ref>. The protocol receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex. | PRosettaC <ref>PRosettaC https://prosettac.weizmann.ac.il</ref> is a computational protocol for the prediction of PROTAC-induced ternary complexes <ref>pmid 32976709</ref>. The protocol receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex. | ||
| + | |||
| + | ==PROTACpedia== | ||
| + | PROTACpedia <ref>PROTACpedia https://protacpedia.weizmann.ac.il/</ref> is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs). | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 08:00, 28 February 2021
PROTAC (proteolysis-targeting chimera) is a small protein with two active domains joined by a linker.
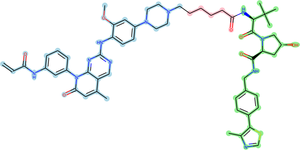
Representative PROTAC from PROTACpedia targeting EGFR Epidermal growth factor receptor P00533. target binding ligandlinkerE3 binder
Predicting PROTACs
PRosettaC [1] is a computational protocol for the prediction of PROTAC-induced ternary complexes [2]. The protocol receives as input, two protein structures (protein target and E3 ligase), including their appropriate ligands (binders), as well as the PROTAC chemical structure in a SMILES representation, and outputs predicted models for the ternary complex.
PROTACpedia
PROTACpedia [3] is a collaborative and comprehensive, high-quality and freely accessible resource of manually curated data on Proteolysis Targeting Chimeras (PROTACs).
References
- ↑ PRosettaC https://prosettac.weizmann.ac.il
- ↑ Zaidman D, Prilusky J, London N. PRosettaC: Rosetta based modeling of PROTAC mediated ternary complexes. J Chem Inf Model. 2020 Sep 25. doi: 10.1021/acs.jcim.0c00589. PMID:32976709 doi:http://dx.doi.org/10.1021/acs.jcim.0c00589
- ↑ PROTACpedia https://protacpedia.weizmann.ac.il/
