We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Leanne Price/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
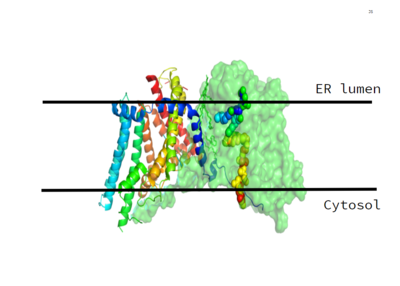
[[Image:Image_1.PNG|400 px|left|thumb|Figure 1. This image shows the location of the DGAT protein within the Endoplasmic Reticulum Membrane]] | [[Image:Image_1.PNG|400 px|left|thumb|Figure 1. This image shows the location of the DGAT protein within the Endoplasmic Reticulum Membrane]] | ||
| - | DGAT, or Diacylglycerol Transferase, makes [https://en.wikipedia.org/wiki/Triglyceride triglycerides] from a [https://en.wikipedia.org/wiki/Diglyceride diglyceride] in plasma. | + | DGAT, or Diacylglycerol Transferase, makes [https://en.wikipedia.org/wiki/Triglyceride triglycerides] from a [https://en.wikipedia.org/wiki/Diglyceride diglyceride] in plasma. DGAT is a polytopic endoplasmic reticulum membrane protein embedded within the membrane of the ER. DGAT is highly expressed in epithelial cells of the small intenstine. It can also be found in the liver, where it helps synthesize fats for storage, and the female mammary glands, where it produces fat in the milk. |
==Function== | ==Function== | ||
Revision as of 19:57, 16 March 2021
DGAT Human
| |||||||||||
References
Wang, L, Qian, H, Nian, Y, Han, Y, Ren, Z, Hanzhi, Z, Hu, L, Prasad, B.V., Laganowsky, A, Yan, N, Zhou, M. (2020). Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature, 581, 329-332.
Sui, X, Wang, K, Gluchowski, N, Elliott, S, Liao, M, Walther, T, Farese Jr, R. (2020). Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature, 581, 323-328.
Rodriguez, A. M. (2020). Zooming in on the architecture of DGAT1, a transmembrane lipid factory. Baylor College of Medicine.
Stone, S. J. Mammalian Diacylglycerol Acyltransferases (DGAT). AOCS Lipid Library.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Student Contributors
- Justin Smith
- Eloi Bigirimana
- Leanne Price