We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2hue
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='2hue' size='340' side='right'caption='[[2hue]], [[Resolution|resolution]] 1.70Å' scene=''> | <StructureSection load='2hue' size='340' side='right'caption='[[2hue]], [[Resolution|resolution]] 1.70Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
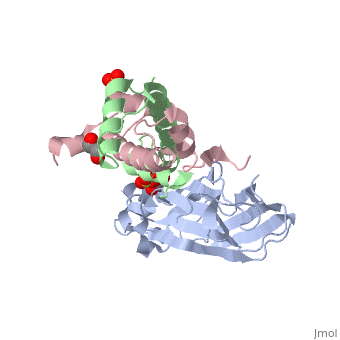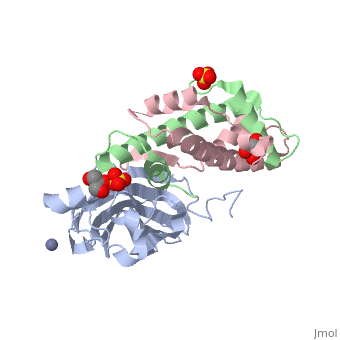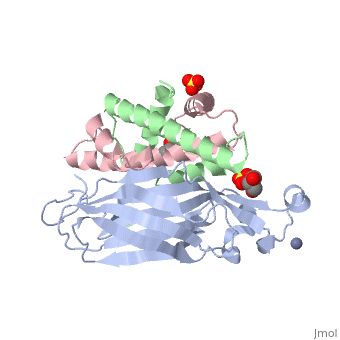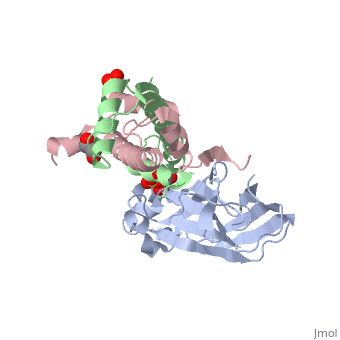
| - | <table><tr><td colspan='2'>[[2hue]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[2hue]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae] and [https://en.wikipedia.org/wiki/Xenopus_laevis Xenopus laevis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2HUE OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2HUE FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.7Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> |
| - | < | + | |
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2hue FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2hue OCA], [https://pdbe.org/2hue PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2hue RCSB], [https://www.ebi.ac.uk/pdbsum/2hue PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2hue ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2hue FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2hue OCA], [https://pdbe.org/2hue PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2hue RCSB], [https://www.ebi.ac.uk/pdbsum/2hue PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2hue ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/ASF1_YEAST ASF1_YEAST] Histone chaperone that facilitates histone deposition and histone exchange and removal during nucleosome assembly and disassembly. Facilitates histone deposition through both replication-dependent and replication-independent chromatin assembly pathways. Cooperates with chromatin assembly factor 1 (CAF-1) to promote replication-dependent chromatin assembly and with the HIR complex to promote replication-independent chromatin assembly, which may occur during transcription and DNA repair. May be required for the maintenance of a subset of replication elongation factors, including DNA polymerase epsilon, the RFC complex and PCNA, at stalled replication forks. Also required for acetylation of histone H3 on 'Lys-9' and 'Lys-56'.<ref>PMID:9290207</ref> <ref>PMID:10591219</ref> <ref>PMID:11412995</ref> <ref>PMID:11331602</ref> <ref>PMID:11731479</ref> <ref>PMID:11731480</ref> <ref>PMID:11404324</ref> <ref>PMID:11172707</ref> <ref>PMID:11856374</ref> <ref>PMID:11756556</ref> <ref>PMID:12093919</ref> <ref>PMID:14585955</ref> <ref>PMID:15071494</ref> <ref>PMID:15452122</ref> <ref>PMID:15175160</ref> <ref>PMID:15542829</ref> <ref>PMID:15542840</ref> <ref>PMID:15766286</ref> <ref>PMID:16303565</ref> <ref>PMID:15821127</ref> <ref>PMID:15901673</ref> <ref>PMID:16020781</ref> <ref>PMID:16143623</ref> <ref>PMID:16039596</ref> <ref>PMID:15632066</ref> <ref>PMID:15891116</ref> <ref>PMID:16141196</ref> <ref>PMID:15840725</ref> <ref>PMID:16815704</ref> <ref>PMID:16936140</ref> <ref>PMID:16582440</ref> <ref>PMID:16407267</ref> <ref>PMID:17046836</ref> <ref>PMID:16678113</ref> <ref>PMID:16501045</ref> <ref>PMID:16627621</ref> <ref>PMID:17107956</ref> <ref>PMID:17320445</ref> <ref>PMID:14680630</ref> | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 21: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2hue ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2hue ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | Anti-silencing function 1 (Asf1) is a highly conserved chaperone of histones H3/H4 that assembles or disassembles chromatin during transcription, replication, and repair. The structure of the globular domain of Asf1 bound to H3/H4 determined by X-ray crystallography to a resolution of 1.7 Angstroms shows how Asf1 binds the H3/H4 heterodimer, enveloping the C terminus of histone H3 and physically blocking formation of the H3/H4 heterotetramer. Unexpectedly, the C terminus of histone H4 that forms a mini-beta sheet with histone H2A in the nucleosome undergoes a major conformational change upon binding to Asf1 and adds a beta strand to the Asf1 beta sheet sandwich. Interactions with both H3 and H4 were required for Asf1 histone chaperone function in vivo and in vitro. The Asf1-H3/H4 structure suggests a "strand-capture" mechanism whereby the H4 tail acts as a lever to facilitate chromatin disassembly/assembly that may be used ubiquitously by histone chaperones. | ||
| - | |||
| - | Structural basis for the histone chaperone activity of Asf1.,English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK Cell. 2006 Nov 3;127(3):495-508. PMID:17081973<ref>PMID:17081973</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 2hue" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| Line 39: | Line 29: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: African clawed frog]] | ||
| - | [[Category: Atcc 18824]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: | + | [[Category: Saccharomyces cerevisiae]] |
| - | [[Category: | + | [[Category: Xenopus laevis]] |
| - | [[Category: | + | [[Category: Churchill MEA]] |
| - | [[Category: | + | [[Category: English CM]] |
| - | [[Category: | + | [[Category: Tyler JK]] |
| - | + | ||
Current revision
Structure of the H3-H4 chaperone Asf1 bound to histones H3 and H4
| |||||||||||