User:Madison Unger/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
An important residue in the ACAT active site is His460, a [http://en.wikipedia.org/wiki/Histidine Histidine], which is located in the middle of the tunnels. It is thought that His460 is located on TM7 (Qian et al.). When converting to a cholesteryl ester, the <scene name='87/877507/His460/5'>His460</scene> acts as a catalytic base that deprotonates the cholesterol. An [http://en.wikipedia.org/wiki/Asparagine asparagine] Asn421 is another important residue in the reaction that is able to form a [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] with acyl-CoA for stabilization. | An important residue in the ACAT active site is His460, a [http://en.wikipedia.org/wiki/Histidine Histidine], which is located in the middle of the tunnels. It is thought that His460 is located on TM7 (Qian et al.). When converting to a cholesteryl ester, the <scene name='87/877507/His460/5'>His460</scene> acts as a catalytic base that deprotonates the cholesterol. An [http://en.wikipedia.org/wiki/Asparagine asparagine] Asn421 is another important residue in the reaction that is able to form a [http://en.wikipedia.org/wiki/Hydrogen_bond hydrogen bond] with acyl-CoA for stabilization. | ||
=== Important Interactions === | === Important Interactions === | ||
| - | Between the two protomers in each dimer, [http://en.wikipedia.org/wiki/Van_der_Waals_force | + | Between the two protomers in each dimer, [http://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals] interactions occur between TM1 of one [http://en.wikipedia.org/wiki/Protomer protomer] and the lumenal TM6 and the cytosolic TM9 of the other protomer. The two dimers make limited contact within the membrane through an interface that is thought to be located between TM2, TM5, TM6 and IH2. |
== Proposed Mechanism == | == Proposed Mechanism == | ||
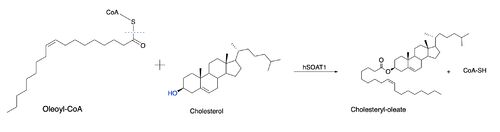
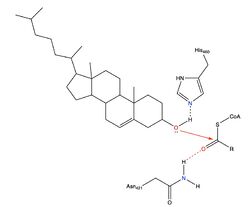
Due to limited high-resolution structural representations of ACAT, its mechanism remains ambiguous. However, the general mechanism involving the substrates and products of ACAT is understood<ref>PMID:32424158</ref>. [[Image:Thenewacatmech1.jpg|500px|left|thumb|Figure 2: Proposed mechanism for ACAT by Guan et al.]] In this reaction, [http://en.wikipedia.org/wiki/Stearoyl-CoA_9-desaturase oleoyl-CoA] and cholesterol are the reactants and they undergo the reaction catalyzed by ACAT to form cholesteryl-oleate which is used as a storage form of cholesterol. The hydroxyl group on cholesterol is deprotonated, then attacks the [http://en.wikipedia.org/wiki/Thioester thioester] bond of oleoyl-CoA, kicking off CoA-SH as a leaving group. This mechanism is shown in Figure 2. [[Image:Thenewacatmech2.jpg|250px|right|thumb|Figure 3: Mechanism for ACAT proposed by Qian et al.]] | Due to limited high-resolution structural representations of ACAT, its mechanism remains ambiguous. However, the general mechanism involving the substrates and products of ACAT is understood<ref>PMID:32424158</ref>. [[Image:Thenewacatmech1.jpg|500px|left|thumb|Figure 2: Proposed mechanism for ACAT by Guan et al.]] In this reaction, [http://en.wikipedia.org/wiki/Stearoyl-CoA_9-desaturase oleoyl-CoA] and cholesterol are the reactants and they undergo the reaction catalyzed by ACAT to form cholesteryl-oleate which is used as a storage form of cholesterol. The hydroxyl group on cholesterol is deprotonated, then attacks the [http://en.wikipedia.org/wiki/Thioester thioester] bond of oleoyl-CoA, kicking off CoA-SH as a leaving group. This mechanism is shown in Figure 2. [[Image:Thenewacatmech2.jpg|250px|right|thumb|Figure 3: Mechanism for ACAT proposed by Qian et al.]] | ||
Revision as of 19:57, 29 March 2021
ACAT/SOAT
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ Rogers MA, Liu J, Song BL, Li BL, Chang CC, Chang TY. Acyl-CoA:cholesterol acyltransferases (ACATs/SOATs): Enzymes with multiple sterols as substrates and as activators. J Steroid Biochem Mol Biol. 2015 Jul;151:102-7. doi: 10.1016/j.jsbmb.2014.09.008., Epub 2014 Sep 12. PMID:25218443 doi:http://dx.doi.org/10.1016/j.jsbmb.2014.09.008
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Leah Goehring
- Gabby Smith
- Anna Campbell