We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Leaman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
== Structural highlights == | == Structural highlights == | ||
| + | |||
| + | == Conformation == | ||
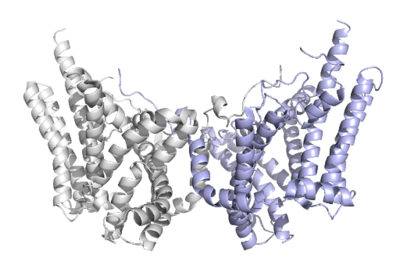
| + | There are three domains within the DGAT enzyme, cytosolic, transmembrane, and lumenal. Most of the enzyme exists within the membrane with small portions peeking out into the cytosol of the cell or lumen in the surrounding tissue. DGAT exists as a homodimer of two identical chains, A and B. The homodimer interface is stabilized in two different ways. First, the transmembrane region is stabilized through large <scene name='87/877557/Hydrophobic_interface/1'>hydrophobic interactions</scene> between the TM1 helices on each of the chains. The second is through extensive <scene name='87/877557/Hydrophilic_interface/1'>hydrogen bonding interactions</scene> between the two chains in the cytosolic domain. | ||
| + | |||
=== Active Site === | === Active Site === | ||
| + | The active site of DGAT is in the transmembrane region of the enzyme. When it is in its <scene name='87/877557/His_no_oleoyl/3'>unbound</scene> state and no oleoyl-CoA is in its tunnel, Met434 hydrogen bonds to the catalytic Histidine, His415, which stabilizes the conformation. There are no major conformation changes that take place upon oleoyl-CoA binding into the cytosolic tunnel, however, several key residues change conformation to allow for the entrance of the ligand. In its <scene name='87/877557/Active_site/6'>bound</scene> conformation, His415 hydrogen bonds to Gln465 which stabilizes the Histidine and allows it to be positioned near the thioester bond of the oleoyl-CoA | ||
| + | |||
=== Tunnel System === | === Tunnel System === | ||
[[Image:oleoyl coa.png|200 px|right|thumb|Figure 2]]There are two main tunnels that allow the enzymatic activity of DGAT to occur. The first is a cytosolic tunnel where the hydrophilic region of the oleyol-CoA binds to the cytosolic face that forms between helices TM6 and TM7. The CoA region sits at the cytosolic face with the fatty acid chain extending the rest of the way through the enzyme tunnel. When hydrophobic residues were substituted with the standard residues in the cytosolic tunnel, DGAT was completely inactivated. | [[Image:oleoyl coa.png|200 px|right|thumb|Figure 2]]There are two main tunnels that allow the enzymatic activity of DGAT to occur. The first is a cytosolic tunnel where the hydrophilic region of the oleyol-CoA binds to the cytosolic face that forms between helices TM6 and TM7. The CoA region sits at the cytosolic face with the fatty acid chain extending the rest of the way through the enzyme tunnel. When hydrophobic residues were substituted with the standard residues in the cytosolic tunnel, DGAT was completely inactivated. | ||
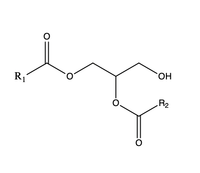
[[Image:DAG.png|200 px|right|thumb|Figure 3]] The second is a <scene name='87/877557/Dag_entrance_tunnel/1'>hydrophobic tunnel</scene> that is orthogonal to the cytosolic tunnel. This tunnel is in the transmembrane region of the enzyme which allows lipids in the membrane to easily access the location. When a molecule of diacylglycerol (DAG), or another acyl acceptor, binds into this hydrophobic tunnel, the catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> the transfer of the acyl group on the bound oleoyl-CoA to the DAG to form a triglyceride. | [[Image:DAG.png|200 px|right|thumb|Figure 3]] The second is a <scene name='87/877557/Dag_entrance_tunnel/1'>hydrophobic tunnel</scene> that is orthogonal to the cytosolic tunnel. This tunnel is in the transmembrane region of the enzyme which allows lipids in the membrane to easily access the location. When a molecule of diacylglycerol (DAG), or another acyl acceptor, binds into this hydrophobic tunnel, the catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> the transfer of the acyl group on the bound oleoyl-CoA to the DAG to form a triglyceride. | ||
| + | |||
=== Mechanism === | === Mechanism === | ||
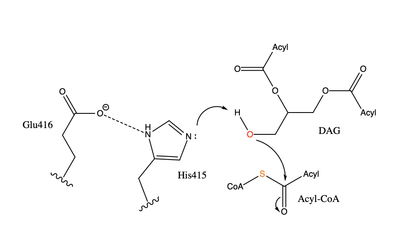
[[Image:dgat chemdraw mechanism.png|400 px|right|thumb|Figure 4]]The catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> deprotonates the hydroxyl group on the C3 of the glycerol backbone. The deprotonated oxygen then makes a nucleophilic attack on the carbonyl carbon of the Acyl-CoA, the electron density gets shifted up to the oxygen and back down to the carbonyl carbon. The sulfur of the Acyl-CoA takes the added electron density and the bond between the sulfur and carbonyl carbon is broken. Glu416 likely provides <scene name='87/878228/His415_and_glu416/2'>Transition State Stabilization</scene> of the His415 despite being outside of the 3 angstrom hydrogen binding distance. This is likely due to the fact that the DGAT 1 enzyme was unable to be visualized with the diacylglycerol in the active site. The entrance and binding of the diacylglycerol may cause conformational changes and shifting of the Glu416 to become closer to the His415. Point mutations made to His415 support the hypothesis that it is essential for stabilization in the active site since the enzyme function was completely eliminated when the mutation was made. | [[Image:dgat chemdraw mechanism.png|400 px|right|thumb|Figure 4]]The catalytic <scene name='87/878228/His415_in_active_site/3'>His415</scene> deprotonates the hydroxyl group on the C3 of the glycerol backbone. The deprotonated oxygen then makes a nucleophilic attack on the carbonyl carbon of the Acyl-CoA, the electron density gets shifted up to the oxygen and back down to the carbonyl carbon. The sulfur of the Acyl-CoA takes the added electron density and the bond between the sulfur and carbonyl carbon is broken. Glu416 likely provides <scene name='87/878228/His415_and_glu416/2'>Transition State Stabilization</scene> of the His415 despite being outside of the 3 angstrom hydrogen binding distance. This is likely due to the fact that the DGAT 1 enzyme was unable to be visualized with the diacylglycerol in the active site. The entrance and binding of the diacylglycerol may cause conformational changes and shifting of the Glu416 to become closer to the His415. Point mutations made to His415 support the hypothesis that it is essential for stabilization in the active site since the enzyme function was completely eliminated when the mutation was made. | ||
Revision as of 21:16, 4 April 2021
Human Diacylglycerol O-Transferase 1
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Megan Leaman
- Grace Hall
- Karina Latsko