User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
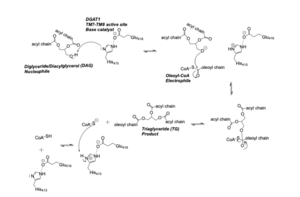
Diacylglycerol acyltransferase (DGAT) is a membrane protein that synthesizes triacylglycerides from its two substrates diacylglycerol (DAG) and fatty acyl-CoA for dietary fat absorption and fat storage. DGAT can be found expressed in the small intestine’s epithelial cells, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk <ref name="Wang">PMID: 32433610</ref>. DGAT is a member of the membrane-bound O- acyltransferases (MBOAT) family <ref name="Sui">PMID: 32433611</ref>. All of the enzymes within this family are transmembrane enzymes that acylate lipids or proteins. Additionally, MBOAT enzymes have a conserved MBOAT core, a channel-like region that acts as the enzyme’s active site. Another feature of note within this MBOAT core is the conserved catalytic Histidine. DGAT is a dimer that has two identical subunits. Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains nine transmembrane alpha-helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. | Diacylglycerol acyltransferase (DGAT) is a membrane protein that synthesizes triacylglycerides from its two substrates diacylglycerol (DAG) and fatty acyl-CoA for dietary fat absorption and fat storage. DGAT can be found expressed in the small intestine’s epithelial cells, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk <ref name="Wang">PMID: 32433610</ref>. DGAT is a member of the membrane-bound O- acyltransferases (MBOAT) family <ref name="Sui">PMID: 32433611</ref>. All of the enzymes within this family are transmembrane enzymes that acylate lipids or proteins. Additionally, MBOAT enzymes have a conserved MBOAT core, a channel-like region that acts as the enzyme’s active site. Another feature of note within this MBOAT core is the conserved catalytic Histidine. DGAT is a dimer that has two identical subunits. Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains nine transmembrane alpha-helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. | ||
| - | |||
== Function == | == Function == | ||
| Line 16: | Line 15: | ||
DGAT is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the dimer interface. Both <scene name='87/877512/Dimer_interface/5'>Hydrogen bonding</scene> and hydrophobic interactions between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together. | DGAT is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the dimer interface. Both <scene name='87/877512/Dimer_interface/5'>Hydrogen bonding</scene> and hydrophobic interactions between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together. | ||
| - | |||
=== Active Site === | === Active Site === | ||
| Line 25: | Line 23: | ||
==== DAG Binding ==== | ==== DAG Binding ==== | ||
DAG enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with the hydrophobic residues Phe342, Leu261, Val381, and Asn378. Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. | DAG enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with the hydrophobic residues Phe342, Leu261, Val381, and Asn378. Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. | ||
| - | |||
==== Acyl-CoA Binding ==== | ==== Acyl-CoA Binding ==== | ||
<scene name='87/877515/Acyl_coa/4'>Acyl-CoA</scene> enters DGAT’s active site through a channel on the cytosolic side of the membrane. In order for the channel to accommodate the fatty acid tail of the Acyl-CoA, His415 must first break its interactions with Met434. This allows for the His415 to swing down and have hydrogen bond interactions with Gln465, thus widening the channel enough for the fatty acid tail to move into the active site. Once within the active site, residues Asn378, Gln437, Met434, His415, and Gln465 directly interact with and stabilize the fatty acid tail within the cytosolic channel of the active site. | <scene name='87/877515/Acyl_coa/4'>Acyl-CoA</scene> enters DGAT’s active site through a channel on the cytosolic side of the membrane. In order for the channel to accommodate the fatty acid tail of the Acyl-CoA, His415 must first break its interactions with Met434. This allows for the His415 to swing down and have hydrogen bond interactions with Gln465, thus widening the channel enough for the fatty acid tail to move into the active site. Once within the active site, residues Asn378, Gln437, Met434, His415, and Gln465 directly interact with and stabilize the fatty acid tail within the cytosolic channel of the active site. | ||
| - | |||
| - | |||
| - | |||
| - | |||
== Disease == | == Disease == | ||
Revision as of 02:34, 5 April 2021
Diacylglycerol acyltransferase, DGAT
| |||||||||||
References
- ↑ 1.0 1.1 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa