User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
==Introduction== | ==Introduction== | ||
| - | Diacylglycerol acyltransferase (<scene name='87/877509/Dgat_overview/2'>DGAT</scene>) is a membrane protein that synthesizes triacylglycerides from its two substrates diacylglycerol (DAG) and fatty acyl-CoA for dietary fat absorption and fat storage. DGAT can be found expressed in the small intestine’s epithelial cells, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk <ref name="Wang">PMID: 32433610</ref>. DGAT is a member of the membrane-bound O- acyltransferases (MBOAT) family <ref name="Sui">PMID: 32433611</ref>. All of the enzymes within this family are transmembrane enzymes that acylate lipids or proteins. | + | Diacylglycerol acyltransferase (<scene name='87/877509/Dgat_overview/2'>DGAT</scene>) is a membrane protein that synthesizes triacylglycerides from its two substrates diacylglycerol (DAG) and fatty acyl-CoA for dietary fat absorption and fat storage. DGAT synthesizes triacylglycerides from diacylglycerol (DAG) and fatty acyl-CoA. DGAT can be found expressed in the small intestine’s epithelial cells, in the liver where it synthesizes fat for storage, and in the female mammary glands where it produces fat for milk <ref name="Wang">PMID: 32433610</ref>. DGAT is a member of the membrane-bound O- acyltransferases (MBOAT) family <ref name="Sui">PMID: 32433611</ref>. All of the enzymes within this family are transmembrane enzymes that acylate lipids or proteins. |
| - | == Function == | ||
| - | |||
| - | DGAT synthesizes triacylglycerides from diacylglycerol (DAG) and fatty acyl-CoA. | ||
== Structure == | == Structure == | ||
[[Image:DGAT_Structure_Drawing.png|900 px|Figure 1: DGAT Structure]] | [[Image:DGAT_Structure_Drawing.png|900 px|Figure 1: DGAT Structure]] | ||
| + | |||
| + | Additionally, MBOAT enzymes have a conserved MBOAT core, a channel-like region that acts as the enzyme’s active site. Another feature of note within this MBOAT core is the conserved catalytic Histidine. DGAT is a dimer that has two identical subunits. Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains nine transmembrane alpha-helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. | ||
| + | |||
| + | |||
=== Dimer Interface === | === Dimer Interface === | ||
DGAT is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the dimer interface. Both <scene name='87/877512/Dimer_interface/5'>Hydrogen bonding</scene> and hydrophobic interactions between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together. | DGAT is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the dimer interface. Both <scene name='87/877512/Dimer_interface/5'>Hydrogen bonding</scene> and hydrophobic interactions between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together. | ||
| - | === | + | ==== Acyl-CoA Binding ==== |
| - | + | <scene name='87/877515/Acyl_coa/4'>Acyl-CoA</scene> enters DGAT’s active site through a channel on the cytosolic side of the membrane. In order for the channel to accommodate the fatty acid tail of the Acyl-CoA, His415 must first break its interactions with Met434. This allows for the His415 to swing down and have hydrogen bond interactions with Gln465, thus widening the channel enough for the fatty acid tail to move into the active site. Once <scene name='87/877509/Surface_active_site/3'>Acyl CoA is bound in the active site</scene>, residues Asn378, Gln437, Met434, His415, and Gln465 directly interact with and stabilize the fatty acid tail within the cytosolic channel of the active site. | |
| - | + | ||
| - | + | ||
==== DAG Binding ==== | ==== DAG Binding ==== | ||
| Line 27: | Line 26: | ||
<scene name='87/877509/Dgat_bound_with_dag/1'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This <scene name='87/877509/Surface_active_site/2'>Active Site Tunnel</scene> or lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with the hydrophobic residues Phe342, Leu261, Val381, and Asn378. Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. | <scene name='87/877509/Dgat_bound_with_dag/1'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This <scene name='87/877509/Surface_active_site/2'>Active Site Tunnel</scene> or lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with the hydrophobic residues Phe342, Leu261, Val381, and Asn378. Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. | ||
| + | === Active Site === | ||
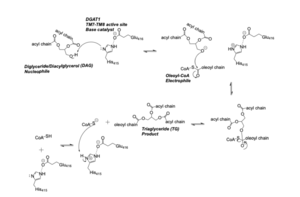
| - | + | The <scene name='87/877509/Active_site/2'>Active Site Residues</scene> are Asn378, Gln437, Gln465, His415, and Met434. The active site of DGAT serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and thus bind the fatty acid to the diacylglycerol. The conserved His415 is able to act catalytically due to a charge relay system, where the neighboring Glu416, due to its negative charge pulls electrons on histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through acyl substitution. The catalytic mechanism for DGAT is shown in Figure 1. | |
| - | <scene name='87/ | + | |
| + | [[Image:DGAT_Mech.png|300 px|right|thumb|Figure 2: DGAT Mechanism]] | ||
| Line 38: | Line 39: | ||
== Relevance == | == Relevance == | ||
| + | |||
<ref name="Wang">PMID: 32433610</ref> | <ref name="Wang">PMID: 32433610</ref> | ||
<ref name="Sui">PMID: 32433611</ref> | <ref name="Sui">PMID: 32433611</ref> | ||
Revision as of 23:07, 5 April 2021
Diacylglycerol acyltransferase, DGAT
| |||||||||||
References
- ↑ 1.0 1.1 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 Denison H, Nilsson C, Lofgren L, Himmelmann A, Martensson G, Knutsson M, Al-Shurbaji A, Tornqvist H, Eriksson JW. Diacylglycerol acyltransferase 1 inhibition with AZD7687 alters lipid handling and hormone secretion in the gut with intolerable side effects: a randomized clinical trial. Diabetes Obes Metab. 2014 Apr;16(4):334-43. doi: 10.1111/dom.12221. Epub 2013 Oct, 31. PMID:24118885 doi:http://dx.doi.org/10.1111/dom.12221
- ↑ 4.0 4.1 Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016 Aug;24(9):1268-73. doi: 10.1038/ejhg.2016.5. Epub 2016 Feb , 17. PMID:26883093 doi:http://dx.doi.org/10.1038/ejhg.2016.5
- ↑ Rebello CJ, Greenway FL. Obesity medications in development. Expert Opin Investig Drugs. 2020 Jan;29(1):63-71. doi:, 10.1080/13543784.2020.1705277. Epub 2019 Dec 19. PMID:31847611 doi:http://dx.doi.org/10.1080/13543784.2020.1705277
- ↑ Scott SA, Mathews TP, Ivanova PT, Lindsley CW, Brown HA. Chemical modulation of glycerolipid signaling and metabolic pathways. Biochim Biophys Acta. 2014 Aug;1841(8):1060-84. doi:, 10.1016/j.bbalip.2014.01.009. Epub 2014 Jan 15. PMID:24440821 doi:http://dx.doi.org/10.1016/j.bbalip.2014.01.009
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa