We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Leanne Price/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
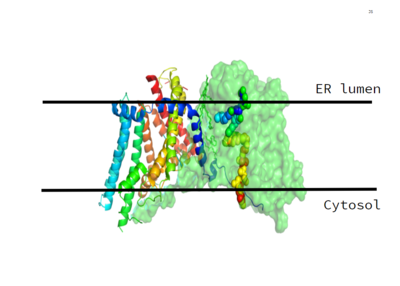
| - | DGAT was originally discovered by its homology to [https://en.wikipedia.org/wiki/Sterol_O-acyltransferase Acyl-CoA cholesterol acyltransferases (ACAT) 1 and 2]. The structure, catalytic mechanism of diacylglycerol acyltransferase, and how DGAT interacts with CoA was discovered using a [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM]. The Cryo-EM map revealed that DGAT forms a dimer, with each subunit containing nine transmembrane helices. The N and C terminals of each | + | DGAT was originally discovered by its homology to [https://en.wikipedia.org/wiki/Sterol_O-acyltransferase Acyl-CoA cholesterol acyltransferases (ACAT) 1 and 2]. The structure, catalytic mechanism of diacylglycerol acyltransferase, and how DGAT interacts with CoA was discovered using a [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM]. The Cryo-EM map revealed that DGAT forms a dimer, with each subunit containing nine transmembrane helices. The N and C terminals of each helix are located on the cytosolic and luminal sides of the endoplasmic reticulum membrane respectively. |
| - | ==Function== | ||
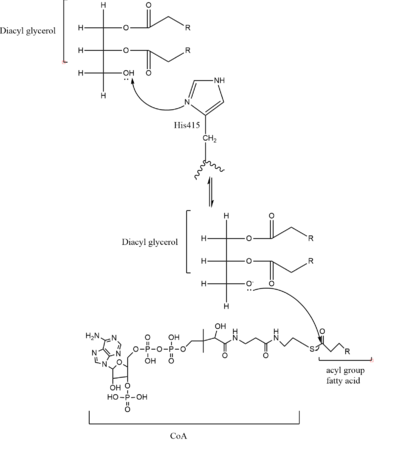
DGAT makes [https://en.wikipedia.org/wiki/Triglyceride triglycerides] from a [https://en.wikipedia.org/wiki/Diglyceride diglyceride] in plasma. In order to do this, DGAT uses two substrates: a fatty acyl-CoA and a diacylglycerol substrate. The basic mechanism consists of a lone pair on a hydroxyl group of glycerol attacking the carbon of the thioester bond of CoA. This results in the breakage of the thioester bond, and the attached acyl group attaches to the glycerol, creating a triglyceride. | DGAT makes [https://en.wikipedia.org/wiki/Triglyceride triglycerides] from a [https://en.wikipedia.org/wiki/Diglyceride diglyceride] in plasma. In order to do this, DGAT uses two substrates: a fatty acyl-CoA and a diacylglycerol substrate. The basic mechanism consists of a lone pair on a hydroxyl group of glycerol attacking the carbon of the thioester bond of CoA. This results in the breakage of the thioester bond, and the attached acyl group attaches to the glycerol, creating a triglyceride. | ||
| - | ==Mutations== | ||
| - | |||
| - | Mutations in the DGAT enzyme are particularly rare. One specific mutation discovered in the DGAT gene leads to congenital diarrhea, electrolyte derangements, protein-losing enteropathy and rickets. This homozygous recessive mutation prevents the expression of the gene and causes the DGAT enzyme to not be expressed. More specifically this mutation has been linked to a 3 base pair deletion at nucleotide position <scene name='87/877601/Maternal_mutation_site/2'>1013 in exon 13</scene>, resulting in an in-frame deletion of a phenylalanine residue at codon 338 maternally, and a C to G transversion at nucleotide position <scene name='87/877601/Paternal_mutation_site/1'>1260 in exon 16</scene>, resulting in a serine to arginine substitution at codon 420 paternally <ref name="Human Protein Atlas" />. | ||
==Structure== | ==Structure== | ||
| Line 46: | Line 42: | ||
As previously mentioned, the Acyl-CoA molecule serves as the leaving group in the DGAT mechanism. This acyl-CoA molecule occupies the cytosolic tunnel, which has a bent architecture. The CoA moiety is at the cytosolic face, while the acyl chain extends through the center towards the endoplasmic reticulum lumen. The distal end of the acyl chain oleoyl-CoA interacts with DGAT deep within the hydrophobic channel, which suggests that the binding of longer acyl chains help accurately position the acyl-donor substrate for the reaction. As the acyl-CoA binds to DGAT, small conformational changes are seen in the active site region, specifically the His415 residue flips towards the endoplasmic reticulum-luminal side when acyl-CoA binds. This conformational change allows a new hydrogen bond to form and positions His415 near the thioester bond of the acyl-CoA. Therefore, the binding of acyl-CoA binding to DGAT results in small, but important, conformational changes in the active site that likely prime the enzyme for catalysis. | As previously mentioned, the Acyl-CoA molecule serves as the leaving group in the DGAT mechanism. This acyl-CoA molecule occupies the cytosolic tunnel, which has a bent architecture. The CoA moiety is at the cytosolic face, while the acyl chain extends through the center towards the endoplasmic reticulum lumen. The distal end of the acyl chain oleoyl-CoA interacts with DGAT deep within the hydrophobic channel, which suggests that the binding of longer acyl chains help accurately position the acyl-donor substrate for the reaction. As the acyl-CoA binds to DGAT, small conformational changes are seen in the active site region, specifically the His415 residue flips towards the endoplasmic reticulum-luminal side when acyl-CoA binds. This conformational change allows a new hydrogen bond to form and positions His415 near the thioester bond of the acyl-CoA. Therefore, the binding of acyl-CoA binding to DGAT results in small, but important, conformational changes in the active site that likely prime the enzyme for catalysis. | ||
| + | ==Mutations== | ||
| + | |||
| + | Mutations in the DGAT enzyme are particularly rare. One specific mutation discovered in the DGAT gene leads to congenital diarrhea, electrolyte derangements, protein-losing enteropathy and rickets. This homozygous recessive mutation prevents the expression of the gene and causes the DGAT enzyme to not be expressed. More specifically this mutation has been linked to a 3 base pair deletion at nucleotide position <scene name='87/877601/Maternal_mutation_site/2'>1013 in exon 13</scene>, resulting in an in-frame deletion of a phenylalanine residue at codon 338 maternally, and a C to G transversion at nucleotide position <scene name='87/877601/Paternal_mutation_site/1'>1260 in exon 16</scene>, resulting in a serine to arginine substitution at codon 420 paternally <ref name="Human Protein Atlas" />. | ||
Revision as of 19:48, 6 April 2021
DGAT Human
| |||||||||||
References
- ↑ 1.0 1.1 https://www.proteinatlas.org/ENSG00000185000-DGAT1/pathology
- ↑ Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
Student Contributors
- Justin Smith
- Eloi Bigirimana
- Leanne Price