User:Josey McKinley/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
== Active Site== | == Active Site== | ||
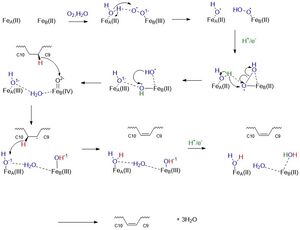
| - | Using X-ray crystallography, two structures of SCD have been found, differing only in their dimetal center. One structure includes the substrate | + | Using X-ray crystallography, two structures of SCD have been found, differing only in their dimetal center. One structure includes the substrate [https://en.wikipedia.org/wiki/Stearoyl-CoA (Stearoyl CoA)] a water molecule, and two <scene name='87/877627/Just_zn_but_zoomed_out/2'>zinc </scene> ions in the center [https://www.rcsb.org/structure/4YMK (4YMK)] (Yonghong). A second structure found more recently includes the product (oleic acid) and two <scene name='87/877627/Zoomed_out_fe/2'>iron</scene> ions in the center [https://www.rcsb.org/structure/6WF2 (6WF2)]. When testing the Zn-centered structure, the enzyme was found to be inactive (Shen). The Zn ions serve as a surrogate for Fe as Zn did not display its typical coordination geometry, tetrahedral; instead, it displayed octahedral geometry which is typical of Fe ion coordination (Yonghong). For the images and links below, the zinc ion-centered structure will be used as it includes the substrate, even though iron is confirmed as the dimetal center. |
| + | |||
The dimetal center is essential to the catalytic activity, as previously demonstrated in the mechanism above. The <scene name='87/877627/Zn_with_measurement/2'>zinc</scene> ions are 6.4 angstroms apart (Yonghong). The ions sit above the kink created by C9 and C10 of the substrate within the active site. The ions are held into the active site through the <scene name='87/877627/His_box_w_o_water/1'>His box</scene> (Kohtaro). The nine coordinating His residues stabilize the ions into the active site forming a non-heme prosthetic group (Kohtaro). The His box is highly conserved among the isoforms of SCD (Shen). | The dimetal center is essential to the catalytic activity, as previously demonstrated in the mechanism above. The <scene name='87/877627/Zn_with_measurement/2'>zinc</scene> ions are 6.4 angstroms apart (Yonghong). The ions sit above the kink created by C9 and C10 of the substrate within the active site. The ions are held into the active site through the <scene name='87/877627/His_box_w_o_water/1'>His box</scene> (Kohtaro). The nine coordinating His residues stabilize the ions into the active site forming a non-heme prosthetic group (Kohtaro). The His box is highly conserved among the isoforms of SCD (Shen). | ||
| + | |||
The <scene name='87/877627/Zn2/2'>ion</scene> closest to C10 of the substrate is 4.7 angstroms away from this carbon (Yonghong). This ion is coordinated by five histidine residues. The <scene name='87/877627/Zn1/2'>ion</scene> closest to C9 of the substrate is 5.2 angstroms away from this carbon (Yonghong). This ion is coordinated with four histidine residues and one water molecule. The <scene name='87/877627/Zn_and_water_round_2/1'>water</scene> is in coordination to the zinc ion closest to it. It occupies the fifth <scene name='87/877627/His_box_w_o_labels/2'>coordination site</scene>. | The <scene name='87/877627/Zn2/2'>ion</scene> closest to C10 of the substrate is 4.7 angstroms away from this carbon (Yonghong). This ion is coordinated by five histidine residues. The <scene name='87/877627/Zn1/2'>ion</scene> closest to C9 of the substrate is 5.2 angstroms away from this carbon (Yonghong). This ion is coordinated with four histidine residues and one water molecule. The <scene name='87/877627/Zn_and_water_round_2/1'>water</scene> is in coordination to the zinc ion closest to it. It occupies the fifth <scene name='87/877627/His_box_w_o_labels/2'>coordination site</scene>. | ||
| + | |||
Residues around the periphery hydrogen bond to the His box to stabilize it. These residues include <scene name='87/877627/D165_correct_one/3'>D165</scene> <scene name='87/877627/E291_correct_one/2'>E291</scene> and <scene name='87/877627/E161_correct_one/2'>E161</scene> (Yonghong). Another residue that stabilizes the active site is <scene name='87/877627/N261_correct_one/3'>N261</scene>. This residue hydrogen bonds to the water molecule (Yonghong). | Residues around the periphery hydrogen bond to the His box to stabilize it. These residues include <scene name='87/877627/D165_correct_one/3'>D165</scene> <scene name='87/877627/E291_correct_one/2'>E291</scene> and <scene name='87/877627/E161_correct_one/2'>E161</scene> (Yonghong). Another residue that stabilizes the active site is <scene name='87/877627/N261_correct_one/3'>N261</scene>. This residue hydrogen bonds to the water molecule (Yonghong). | ||
The His box and periphery residues stabilize the dimetal center and make up the <scene name='87/877627/Active_site_round_3_but_labels/1'>active site</scene> of the enzyme. This allows for the proposed above mechanism to be carried out. | The His box and periphery residues stabilize the dimetal center and make up the <scene name='87/877627/Active_site_round_3_but_labels/1'>active site</scene> of the enzyme. This allows for the proposed above mechanism to be carried out. | ||
Revision as of 02:54, 20 April 2021
Stearoyl-CoA Desaturase 1 from Mus musculus
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Josey McKinley
- Abbey Wells
- Anthony Durand