We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Megan Leaman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
=== Tunnel System === | === Tunnel System === | ||
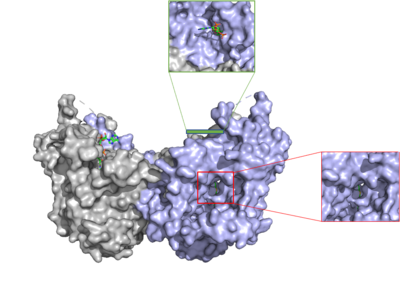
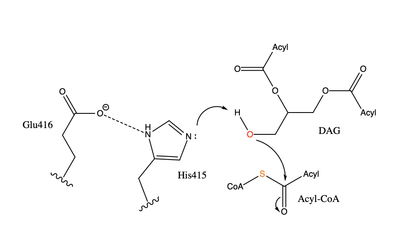
| - | [[Image: | + | [[Image:DGAT tunnels.png|400 px|right|thumb|Figure 3: cytosolic tunnel (green) and hydrophobic tunnel (red)]]There are two main tunnels that allow the enzymatic activity of DGAT to occur. The first is a <scene name='87/877557/Oleoyl_co_a_cytosolic_tunnel/2'>cytosolic tunnel</scene> where the hydrophilic region of the oleyol-CoA binds to the cytosolic face that forms between helices TM6 and TM7 (Fig. 3). The [https://en.wikipedia.org/wiki/Coenzyme_A CoA] region sits at the cytosolic face with the fatty acid chain extending the rest of the way through the enzyme tunnel. <ref name="Wang">PMID:32433610</ref> Researchers are currently unsure which residues are involved in the binding of the oleoyl-coA to the reaction chamber, but they believe that the hydrogen bonding of the coenzyme A motif that sits at the cytosolic face of the tunnel. When hydrophobic residues were substituted with the standard residues in the cytosolic tunnel, DGAT was completely inactivated. <ref name="Wang">PMID:32433610</ref> <ref name="Sui">PMID:32433611</ref> |
The second is a <scene name='87/877557/Dag_entrance_tunnel/4'>hydrophobic tunnel</scene> that is orthogonal to the cytosolic tunnel (Fig. 3). This tunnel is in the transmembrane region of the enzyme which allows lipids in the membrane to easily access the location. <ref name="Sui">PMID:32433611</ref> Researchers have hypothesized that this tunnel is able to differentiate between DAG, its intended substrate, from other groups that would typically interact with an acyl-CoA, like cholesterol, due to its bent architecture. The bent nature of the tunnel would inhibit more stiff and planar molecules from entering the tunnel and interfering with the activity of the enzyme. <ref name="Sui">PMID:32433611</ref> | The second is a <scene name='87/877557/Dag_entrance_tunnel/4'>hydrophobic tunnel</scene> that is orthogonal to the cytosolic tunnel (Fig. 3). This tunnel is in the transmembrane region of the enzyme which allows lipids in the membrane to easily access the location. <ref name="Sui">PMID:32433611</ref> Researchers have hypothesized that this tunnel is able to differentiate between DAG, its intended substrate, from other groups that would typically interact with an acyl-CoA, like cholesterol, due to its bent architecture. The bent nature of the tunnel would inhibit more stiff and planar molecules from entering the tunnel and interfering with the activity of the enzyme. <ref name="Sui">PMID:32433611</ref> | ||
Revision as of 04:23, 23 April 2021
Human Diacylglycerol O-Transferase 1
| |||||||||||
References
- ↑ 1.0 1.1 Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, Erickson SK, Farese RV Jr. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci U S A. 1998 Oct 27;95(22):13018-23. PMID:9789033
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV Jr. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008 Nov;49(11):2283-301. doi: 10.1194/jlr.R800018-JLR200. Epub 2008, Aug 29. PMID:18757836 doi:http://dx.doi.org/10.1194/jlr.R800018-JLR200
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ Haas, J. T., Winter, H. S., Lim, E., Kirby, A., Blumenstiel, B., DeFelice, M., Gabriel, S., Jalas, C., Branski, D., Grueter, C. A., Toporovski, M. S., Walther, T. C., Daly, M. J., & Farese, R. V., Jr (2012). DGAT1 mutation is linked to a congenital diarrheal disorder. The Journal of clinical investigation, 122(12), 4680–4684. https://doi.org/10.1172/JCI64873
- ↑ 6.0 6.1 Gluchowski, N. L., Chitraju, C., Picoraro, J. A., Mejhert, N., Pinto, S., Xin, W., Kamin, D. S., Winter, H. S., Chung, W. K., Walther, T. C., & Farese, R. V., Jr (2017). Identification and characterization of a novel DGAT1 missense mutation associated with congenital diarrhea. Journal of lipid research, 58(6), 1230–1237. https://doi.org/10.1194/jlr.P075119
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Megan Leaman
- Grace Hall
- Karina Latsko