User:Kaitlyn Roberts/Sandbox 2
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
== Structure == | == Structure == | ||
=== Tertiary Structure === | === Tertiary Structure === | ||
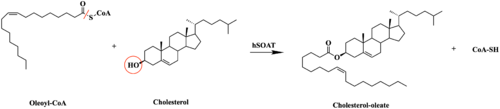
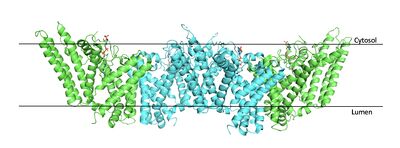
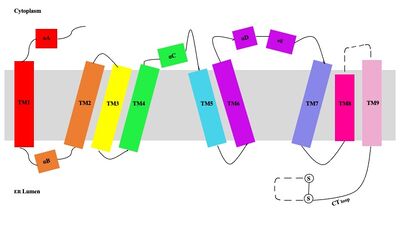
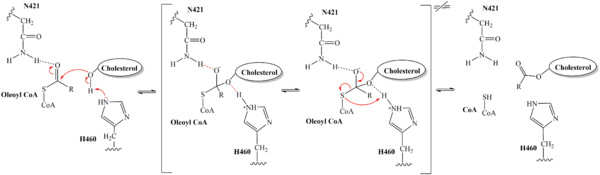
| - | [[Image:Tetramerlabels.jpeg|400 px|right|thumb|'''Figure 2.''' Tetramer unit of SOAT shown as it sits within the membrane. Each dimer is composed of a green and blue chain with the corresponding monomer chains colored the same. [http://www.rcsb.org/structure/6P2P PBD 6P2P]]] The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> tetramer structure or a <scene name='87/877559/Tetramer/11'>dimer of dimers</scene>. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene>which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> structures. The hydrophobic interactions between the residues on the surface of the dimers rely on the structure and orientation of the residues and mutating them will lead to the breakup of the dimer into the monomers. The importance of the dimeric structure is shown as the enzyme was unable to properly function after the reduction to the monomer units. <ref name="Guan" /> Each monomer is made up of nine <scene name='87/877559/Helices_1-9/4'>transmembrane helices</scene> that can be labeled TM1 through TM9. [[Image:Helicesdiagram1.jpeg|400 px|right|thumb|'''Figure 3.''' Labeled helices of SOAT within the membrane]] Substantial [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] help to maintain this dimeric structure at the TM1 helice in one of the monomers and the TM6 [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] and the TM9 [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] in the other monomer. The essential helices from the two monomers (TM1, TM5, TM6, and TM9) helps to form the tunnels and catalytic active site, as well as to help the efforts to stabilize the monomer interactions in the dimer. <ref name="Qian">PMID:32433614</ref> | + | [[Image:Tetramerlabels.jpeg|400 px|right|thumb|'''Figure 2.''' Tetramer unit of SOAT shown as it sits within the membrane. Each dimer is composed of a green and blue chain with the corresponding monomer chains colored the same. [http://www.rcsb.org/structure/6P2P PBD 6P2P]]] The overall structure of the enzyme is a <scene name='87/877559/Tetramer/10'>tetramer</scene> tetramer structure or a <scene name='87/877559/Tetramer/11'>dimer of dimers</scene>. The functional building block of SOAT is a <scene name='87/877559/Dimer/3'>dimer</scene> which is made up of two identical <scene name='87/877559/Monomer/5'>monomer</scene> structures. The hydrophobic interactions between the residues on the surface of the dimers rely on the structure and orientation of the residues and mutating them will lead to the breakup of the dimer into the monomers. The importance of the dimeric structure is shown as the enzyme was unable to properly function after the reduction to the monomer units. <ref name="Guan" /> Each monomer is made up of nine <scene name='87/877559/Helices_1-9/4'>transmembrane helices</scene> that can be labeled TM1 through TM9. [[Image:Helicesdiagram1.jpeg|400 px|right|thumb|'''Figure 3.''' Labeled helices of SOAT within the membrane]] Substantial [https://en.wikipedia.org/wiki/Van_der_Waals_force van der Waals interactions] help to maintain this dimeric structure at the TM1 helice in one of the monomers and the TM6 [https://en.wikipedia.org/wiki/Lumen_(anatomy) lumenal segment] and the TM9 [https://en.wikipedia.org/wiki/Cytosol cytosolic segment] in the other monomer. The essential helices from the two monomers (TM1, TM5, TM6, and TM9) helps to form the tunnels and catalytic active site, as well as to help the efforts to stabilize the monomer interactions in the dimer. <ref name="Qian">PMID:32433614</ref> |
=== Tunnel System === | === Tunnel System === | ||
Revision as of 02:57, 26 April 2021
Human Sterol O-acyltransferase
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 Guan C, Niu Y, Chen SC, Kang Y, Wu JX, Nishi K, Chang CCY, Chang TY, Luo T, Chen L. Structural insights into the inhibition mechanism of human sterol O-acyltransferase 1 by a competitive inhibitor. Nat Commun. 2020 May 18;11(1):2478. doi: 10.1038/s41467-020-16288-4. PMID:32424158 doi:http://dx.doi.org/10.1038/s41467-020-16288-4
- ↑ 2.0 2.1 2.2 2.3 Qian H, Zhao X, Yan R, Yao X, Gao S, Sun X, Du X, Yang H, Wong CCL, Yan N. Structural basis for catalysis and substrate specificity of human ACAT1. Nature. 2020 May;581(7808):333-338. doi: 10.1038/s41586-020-2290-0. Epub 2020 May, 13. PMID:32433614 doi:http://dx.doi.org/10.1038/s41586-020-2290-0
- ↑ 3.0 3.1 Bhattacharyya R, Kovacs DM. ACAT inhibition and amyloid beta reduction. Biochim Biophys Acta. 2010 Aug;1801(8):960-5. doi: 10.1016/j.bbalip.2010.04.003. , Epub 2010 Apr 14. PMID:20398792 doi:http://dx.doi.org/10.1016/j.bbalip.2010.04.003
- ↑ 4.0 4.1 Huttunen HJ, Kovacs DM. ACAT as a drug target for Alzheimer's disease. Neurodegener Dis. 2008;5(3-4):212-4. doi: 10.1159/000113705. Epub 2008 Mar 6. PMID:18322393 doi:http://dx.doi.org/10.1159/000113705
- ↑ Chang C, Dong R, Miyazaki A, Sakashita N, Zhang Y, Liu J, Guo M, Li BL, Chang TY. Human acyl-CoA:cholesterol acyltransferase (ACAT) and its potential as a target for pharmaceutical intervention against atherosclerosis. Acta Biochim Biophys Sin (Shanghai). 2006 Mar;38(3):151-6. doi:, 10.1111/j.1745-7270.2006.00154.x. PMID:16518538 doi:http://dx.doi.org/10.1111/j.1745-7270.2006.00154.x
- ↑ Ayyagari VN, Wang X, Diaz-Sylvester PL, Groesch K, Brard L. Assessment of acyl-CoA cholesterol acyltransferase (ACAT-1) role in ovarian cancer progression-An in vitro study. PLoS One. 2020 Jan 24;15(1):e0228024. doi: 10.1371/journal.pone.0228024., eCollection 2020. PMID:31978092 doi:http://dx.doi.org/10.1371/journal.pone.0228024
Student Contributors
- Kylie Pfeifer
- Stephanie Pellegrino
- Kaitlyn Roberts