We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Overview=== | ===Overview=== | ||
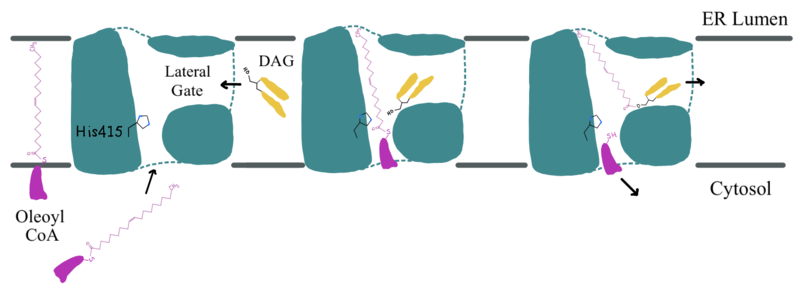
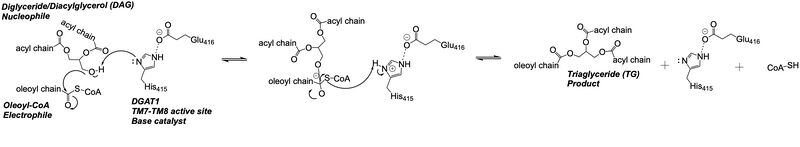
| - | DGAT1 is a dimer that has two identical <scene name='87/877512/Labeled_subunits/1'>subunits</scene>. Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains <scene name='87/877512/Labeled_helices/2'>nine transmembrane helices</scene> (TM), 2 intracellular loops (IL), and one ER lumenal loop (EL). TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. A schematic of DGAT1’s structure is shown in Figure 1. | + | DGAT1 is a dimer that has two identical <scene name='87/877512/Labeled_subunits/1'>subunits</scene>. Each of the individual subunits contains an MBOAT core that acts as its active site. Each subunit also contains <scene name='87/877512/Labeled_helices/2'>nine transmembrane helices</scene> (TM), 2 intracellular loops (IL), and one ER lumenal loop (EL) <ref name="Wang">PMID: 32433610</ref>. TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. A schematic of DGAT1’s structure is shown in Figure 1. |
=== Dimer Interface === | === Dimer Interface === | ||
| - | DGAT1 is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the <scene name='87/877512/Dimer-interface/1'>dimer interface</scene>. Both <scene name='87/877512/Dimer_interface/6'>hydrogen bonding and hydrophobic interactions</scene> between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together. | + | DGAT1 is a dimer that has two identical subunits with 9 transmembrane alpha helices (TM), 2 intracellular loops (IL), and one ER luminal loop (EL). The dimer is held together at the <scene name='87/877512/Dimer-interface/1'>dimer interface</scene>. Both <scene name='87/877512/Dimer_interface/6'>hydrogen bonding and hydrophobic interactions</scene> between the residues of the TM1 and EL1 regions of both subunits act to hold the subunits of the dimer together <ref name="Sui">PMID: 32433611</ref>. |
| Line 60: | Line 60: | ||
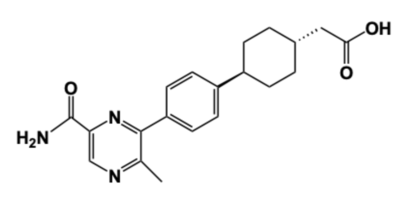
DGAT1 is an important enzyme in the synthesis of triacylglycerides and has relevance in the research of diseases that involve triacylglyceride accumulation, like obesity and NAFLD, or triacylglyceride reduction, like congenital PLE. There is an opportunity within pharmaceuticals for developing DGAT1 inhibitors like AZD7687 to lessen the severity of various diseases through a decrease in triacylglyceride storage. However, inhibitors will have to work around the symptoms of triacylglyceride reduction, as seen through the impact of the Leu295Pro mutation in congenital PLE patients. Developing an inhibitor that can balance DGAT1 efficacy between excess and deprivation of triacylglyceride synthesis can improve treatments for obesity, NAFLD, and other triacylglyceride storage diseases. | DGAT1 is an important enzyme in the synthesis of triacylglycerides and has relevance in the research of diseases that involve triacylglyceride accumulation, like obesity and NAFLD, or triacylglyceride reduction, like congenital PLE. There is an opportunity within pharmaceuticals for developing DGAT1 inhibitors like AZD7687 to lessen the severity of various diseases through a decrease in triacylglyceride storage. However, inhibitors will have to work around the symptoms of triacylglyceride reduction, as seen through the impact of the Leu295Pro mutation in congenital PLE patients. Developing an inhibitor that can balance DGAT1 efficacy between excess and deprivation of triacylglyceride synthesis can improve treatments for obesity, NAFLD, and other triacylglyceride storage diseases. | ||
| + | |||
| + | <ref name="Wang">PMID: 32433610</ref> | ||
| + | <ref name="Sui">PMID: 32433611</ref> | ||
| + | <ref name="Stephen">PMID: 26883093</ref> | ||
| + | <ref name="Denison">PMID: 24118885</ref> | ||
| + | <ref name=”Ma”>PMID: 30283133</ref> | ||
Revision as of 19:32, 26 April 2021
Diacylglycerol acyltransferase 1, DGAT1, synthesizes triacylglycerides
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 2.2 2.3 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 3.2 Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018 Oct;562(7726):286-290. doi: 10.1038/s41586-018-0568-2. Epub 2018 Oct, 3. PMID:30283133 doi:http://dx.doi.org/10.1038/s41586-018-0568-2
- ↑ 4.0 4.1 4.2 4.3 Denison H, Nilsson C, Lofgren L, Himmelmann A, Martensson G, Knutsson M, Al-Shurbaji A, Tornqvist H, Eriksson JW. Diacylglycerol acyltransferase 1 inhibition with AZD7687 alters lipid handling and hormone secretion in the gut with intolerable side effects: a randomized clinical trial. Diabetes Obes Metab. 2014 Apr;16(4):334-43. doi: 10.1111/dom.12221. Epub 2013 Oct, 31. PMID:24118885 doi:http://dx.doi.org/10.1111/dom.12221
- ↑ 5.0 5.1 Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016 Aug;24(9):1268-73. doi: 10.1038/ejhg.2016.5. Epub 2016 Feb , 17. PMID:26883093 doi:http://dx.doi.org/10.1038/ejhg.2016.5
- ↑ Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018 Oct;562(7726):286-290. doi: 10.1038/s41586-018-0568-2. Epub 2018 Oct, 3. PMID:30283133 doi:http://dx.doi.org/10.1038/s41586-018-0568-2
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa