This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:Jacob Holt/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 44: | Line 44: | ||
=== Ligand Binding Pocket and Histidine Coordination === | === Ligand Binding Pocket and Histidine Coordination === | ||
| - | The ligand binding pocket is a narrow tunnel that extends approximately 24 Å into the mostly hydrophobic interior of the protein. The ligand is stabilized by bending into a kinked conformation which creates a tight fit in the binding pocket tunnel, and by a hydrogen bond that occurs between the <scene name='87/877552/W258/ | + | The ligand binding pocket is a narrow tunnel that extends approximately 24 Å into the mostly hydrophobic interior of the protein. The ligand is stabilized by bending into a kinked conformation which creates a tight fit in the binding pocket tunnel, and by a hydrogen bond that occurs between the <scene name='87/877552/W258/4'>W 258</scene> side chain and the acyl carbonyl<ref name="Bai" />. The kink in the tunnel is formed by the conserved residues, <scene name='87/877552/Desaturation_site/10'>T 257 and W 149</scene> which are stabilized by the hydrogen bond shared with Q143<ref name="Bai" />. There are <scene name='87/877552/Substrate_orientation_w_fe/7'>two Fe2+ ions</scene> that interact with the substrate; the Fe2+ ions are coordinated by <scene name='87/877552/Histidine_coordination/8'>9 invariant histidine residues</scene>. <scene name='87/877552/Substrate_oreintation_fe_90deg/4'>When rotated 90 degrees</scene> the ligand is seen to be in a eclipsed position, indicating it is in its post-reaction form. One metal ion is coordinated by 4 histidines residues and a water molecule, and the other metal ion is coordinated by 5 histidine residues<ref name="Bai" />. The histidine residues position the metal ions 6.4 Å apart<ref name="Bai" />. |
=== Desaturation Site === | === Desaturation Site === | ||
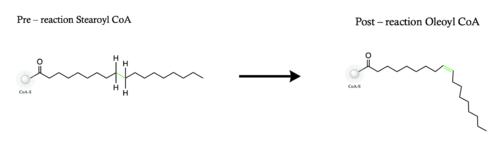
| - | The ligand is desaturated at carbons 9 and 10<ref name="Shen" />. The desaturation site of the ligand takes place inside the active site tunnel which enforces correct positioning of the substrate<ref name="Bai" />. Before the reaction occurs, the ligand is in a gauche conformation at the desaturation site. This was determined by accidental usage of <scene name='87/877552/Pre_reaction_substrate_zn/ | + | The ligand is desaturated at carbons 9 and 10<ref name="Shen" />. The desaturation site of the ligand takes place inside the active site tunnel which enforces correct positioning of the substrate<ref name="Bai" />. Before the reaction occurs, the ligand is in a gauche conformation at the desaturation site. This was determined by accidental usage of <scene name='87/877552/Pre_reaction_substrate_zn/3'>Zn+ ions</scene> which allowed for binding of the substrate but prevented the reaction<ref name="Shen" />. The product is in a cis conformation post-reaction. The product structure was determined using Fe2+ metal ions which allowed for the full reaction to take place<ref name="Shen" />. The difference between the <scene name='87/877552/Overlay_of_ligands/6'>substrate and product</scene> is the creation of a double bond, and the positioning of carbon 9 and 10 into a eclipsed position |
=== Active Site Cap === | === Active Site Cap === | ||
| - | The two conserved residues of the active site cap are <scene name='87/877552/Active_site_cap/ | + | The two conserved residues of the active site cap are <scene name='87/877552/Active_site_cap/8'>Y 104 and G 287</scene>. These two residues form a hydrogen bond creating a rigid barrier at the end of the active site to keep the ligand from moving during the reaction<ref name="Bai" />. The active site cap is also used in determining the substrate length when entering the active site<ref name="Bai" />. |
=== Catalytic Molecule === | === Catalytic Molecule === | ||
| - | The <scene name='87/877552/Water/ | + | The <scene name='87/877552/Water/4'>catalytic molecule</scene> of the SCD1 enzyme is a water molecule, coordinated by <scene name='87/877552/Asparagine_h20_stabilization/3'>N 261</scene> via hydrogen bonding<ref name="Bai" />. The water molecule is 2.2 Å away from the Fe2+ metal ion molecule<ref name="Bai" />. It interacts with the Fe2+ ion to make highly reactive radicals that are able to desaturate the highly stable carbon chain<ref name="Yu">DOI:10.1021/acscatal.9b00456 </ref>. It is through the coordination of these ions by the histidines that the <scene name='87/877552/Diiron_center/9'>substrate and catalytic molecule</scene> are able to be positioned within the vicinity of carbons 9 and 10 of the ligand<ref name="Yu" />. |
=== Substrate Entering and Leaving === | === Substrate Entering and Leaving === | ||
| - | The substrate enters the active site through the active site tunnel and undergoes a <scene name='87/877552/Substrate_entry_exit/ | + | The substrate enters the active site through the active site tunnel and undergoes a <scene name='87/877552/Substrate_entry_exit/4'>conformational change</scene> to conform to the kinked shape of the tunnel<ref name="Bai" />. Upon the substrate being converted to its final form, the product is laterally released from the protein due to a lateral egress of H1 and H2 caused by the deformation in the hydrogen bonding of the residues N 143 and T 257<ref name="Bai" />. |
| Line 70: | Line 70: | ||
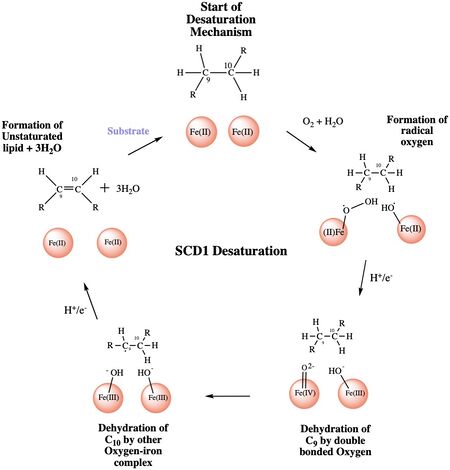
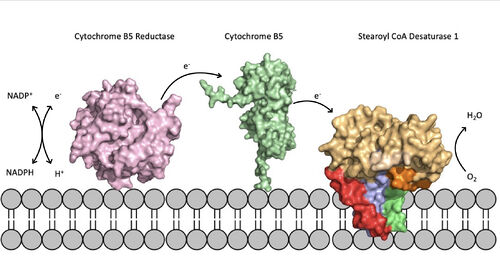
'''Step 1''': Addition of O2 and H2O which react with the Fe2+ ions to create oxygen radicals on the iron ions (Figure 4). | '''Step 1''': Addition of O2 and H2O which react with the Fe2+ ions to create oxygen radicals on the iron ions (Figure 4). | ||
| - | '''Step 2''': Electron/proton pair is brought in via the electron transport chain; this increases the oxidation of both iron ions, gets rid of the radicals, and creates an active Fe-oxyl molecule (Figure). | + | '''Step 2''': Electron/proton pair is brought in via the electron transport chain; this increases the oxidation of both iron ions, gets rid of the radicals, and creates an active Fe-oxyl molecule (Figure)<ref name="Yu" />. Fe-oxyl molecule is reactive enough, due to the change in the oxidation state, to pull off the first hydrogen on carbon 9 (Figure 4). |
| - | '''Step 3''': An unstable radical intermediate of the 18-carbon acyl-CoA ligand is formed which reacts with the other Fe-O molecule, in the +3 state, to pull of the second hydrogen and form the final product ( | + | '''Step 3''': An unstable radical intermediate of the 18-carbon acyl-CoA ligand is formed which reacts with the other Fe-O molecule, in the +3 state, to pull of the second hydrogen and form the final product (Figure 4)<ref name="Yu" />. |
'''Step 4''': Another electron/proton pair is brought in to create three H2O molecules and to take the Fe ions back down to their original oxidation state of +2 (Figure 4)<ref name="Yu" />. | '''Step 4''': Another electron/proton pair is brought in to create three H2O molecules and to take the Fe ions back down to their original oxidation state of +2 (Figure 4)<ref name="Yu" />. | ||
Revision as of 14:45, 27 April 2021
Desaturation of Fatty Acids using Stearoyl-CoA Desaturase-1 Enzyme
| |||||||||||
Student Contributions
Carson Maris, Jess Kersey, Jacob Holt