We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Sarah Maarouf/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 31: | Line 31: | ||
=== N-terminal α/β-hydrolase domain of LPL === | === N-terminal α/β-hydrolase domain of LPL === | ||
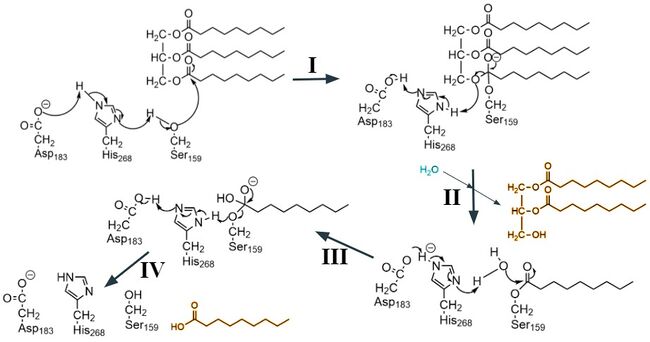
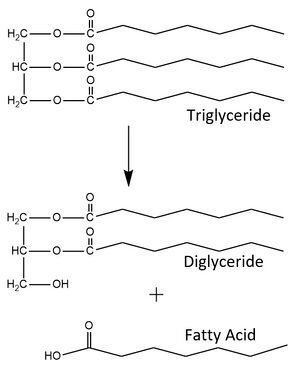
| - | The <scene name='87/877554/N-terminal_domain/4'>N-terminal domain</scene> is composed of one antiparallel β-sheet and seven parallel β-sheets that are located between five α-helices. This is the domain responsible for hydrolysis of lipid substrates, as it contains the <scene name='87/877554/Active_site_residues/11'>catalytic triad</scene> and houses the <scene name='87/877554/Oxyanion_hole/10'>oxyanion hole</scene> to stabilize the transition state of the substrate. The N-terminal domain includes a <scene name='87/877554/Calcium_ion_coordination_sites/6'>calcium ion that is coordinated by a number of residues</scene> which has been shown to have mutations that may impact LPL enzyme activity. The lid region of the N-terminal domain was imaged in an open conformation, meaning it is not blocking the active site. The <scene name='87/877554/Lid_region/7'>lid region</scene> consists of 2 short α-helices connected by a loop, extending away from the protein. This open conformation allows for many surface-exposed hydrophobic residues (valines,isoleucines, and leucines) to create a hydrophobic patch on the surface of LPL. The lid region helps to control for the entry of lipid substrates into the active site cleft, though specifics about how lipids enter into the active site still | + | The <scene name='87/877554/N-terminal_domain/4'>N-terminal domain</scene> is composed of one antiparallel β-sheet and seven parallel β-sheets that are located between five α-helices. This is the domain responsible for hydrolysis of lipid substrates, as it contains the <scene name='87/877554/Active_site_residues/11'>catalytic triad</scene> and houses the <scene name='87/877554/Oxyanion_hole/10'>oxyanion hole</scene> to stabilize the transition state of the substrate. The N-terminal domain includes a <scene name='87/877554/Calcium_ion_coordination_sites/6'>calcium ion that is coordinated by a number of residues</scene> which has been shown to have mutations that may impact LPL enzyme activity. The lid region of the N-terminal domain was imaged in an open conformation, meaning it is not blocking the active site. The <scene name='87/877554/Lid_region/7'>lid region</scene> consists of 2 short α-helices connected by a loop, extending away from the protein. This open conformation allows for many surface-exposed hydrophobic residues (valines, isoleucines, and leucines) to create a hydrophobic patch on the surface of LPL. The lid region helps to control for the entry of lipid substrates into the active site cleft, though specifics about how lipids enter into the active site still requires further investigation.<ref name="Arora">PMID:31072929</ref> |
=== C-terminal β-barrel domain of LPL === | === C-terminal β-barrel domain of LPL === | ||
Revision as of 14:02, 27 April 2021
H. sapiens Lipoprotein Lipase in complex with GPIHBP1 and triglyceride metabolism
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Fong LG, Young SG, Beigneux AP, Bensadoun A, Oberer M, Jiang H, Ploug M. GPIHBP1 and Plasma Triglyceride Metabolism. Trends Endocrinol Metab. 2016 Jul;27(7):455-469. doi: 10.1016/j.tem.2016.04.013. , Epub 2016 May 14. PMID:27185325 doi:http://dx.doi.org/10.1016/j.tem.2016.04.013
- ↑ 2.0 2.1 Voss CV, Davies BS, Tat S, Gin P, Fong LG, Pelletier C, Mottler CD, Bensadoun A, Beigneux AP, Young SG. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci U S A. 2011 May 10;108(19):7980-4. doi:, 10.1073/pnas.1100992108. Epub 2011 Apr 25. PMID:21518912 doi:http://dx.doi.org/10.1073/pnas.1100992108
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ 4.0 4.1 Young SG, Fong LG, Beigneux AP, Allan CM, He C, Jiang H, Nakajima K, Meiyappan M, Birrane G, Ploug M. GPIHBP1 and Lipoprotein Lipase, Partners in Plasma Triglyceride Metabolism. Cell Metab. 2019 Jul 2;30(1):51-65. doi: 10.1016/j.cmet.2019.05.023. PMID:31269429 doi:http://dx.doi.org/10.1016/j.cmet.2019.05.023
- ↑ Olivecrona G. Role of lipoprotein lipase in lipid metabolism. Curr Opin Lipidol. 2016 Jun;27(3):233-41. doi: 10.1097/MOL.0000000000000297. PMID:27031275 doi:http://dx.doi.org/10.1097/MOL.0000000000000297
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ Hedstrom L. Serine Protease Mechanism and Specificity. Chemical Reviews 2002 102 (12), 4501-4524 https://doi.org/10.1021/cr000033x
Student Contributors
- Aniyah Coles
- Sarah Maarouf
- Audrey Marjamaa
![Figure 2. A cartoon representation of GPIHBP1's N-terminal intrinsically disordered region (IDR) colored green and an surface representation of LPL with electrostatic coloring [acidic (red), neutral (white), basic (blue)] to show how GPIHBP1's N-terminal domain interacts with LPL's basic patch to stabilize LPL structure and activity.](/wiki/images/thumb/7/72/Electro.jpg/250px-Electro.jpg)