We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 69: | Line 69: | ||
[[Image:Photinus_pyralis.jpg|thumb|The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.]] | [[Image:Photinus_pyralis.jpg|thumb|The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.]] | ||
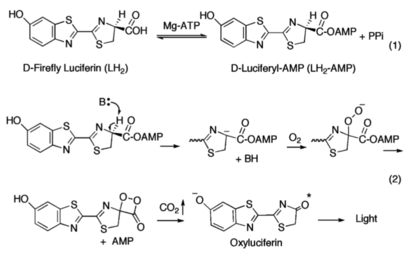
Firefly go glow, super duper cool B~) | Firefly go glow, super duper cool B~) | ||
| + | Firefly luciferase has successfully been shown to act as modulatory bioluminescent indicator in the detection and quantification of protein kinase A activation in living cells <ref name="Newby">Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727</ref> | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 16:09, 27 April 2021
Firefly Luciferase
tttaaarrgggetttt to the right plaaccceee and finishh :(
| |||||||||||
References
- ↑ Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. Biochemistry 38(40), 13223–13230. https://doi.org/10.1021/bi991181o
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. Biochemistry 51(33), 6493-6495. https://doi.org/10.1021/bi300934s
- ↑ Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. Chemical Reviews 97(7), 2651-2674. https://doi.org/10.1021/cr960029e
- ↑ Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. Biochemistry 39(18), 5433-5440. https://doi.org/10.1021/bi9928804
- ↑ Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f
- ↑ Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727