We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Hannah Wright/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
== Structure == | == Structure == | ||
===Overall Structure=== | ===Overall Structure=== | ||
| - | Overall, LPL is a monomer but | + | Overall, LPL is a monomer but in the <scene name='87/877603/Chain_a_and_chain_b/4'>experimental crystal structure</scene> it is paired with another monomer. The two monomers in the experimental structure are identical besides orientation. The two monomers are oriented head to tail. LPL is also complexed with GPIHBP1 (shown as cyan) and is essential for LPL to remain stable and avoid [http://en.wikipedia.org/wiki/Denaturation_(biochemistry) denaturation]. |
===LPL=== | ===LPL=== | ||
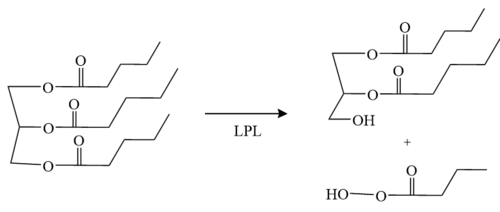
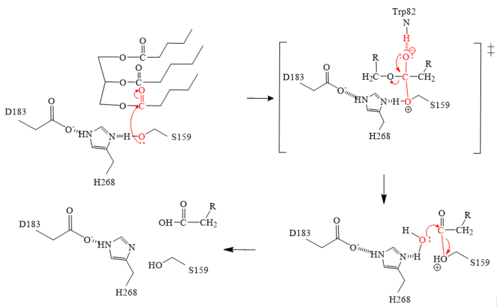
LPL has two main domains, the larger N-terminus domain containing the active site and the smaller C-terminus domain. These two domains are connected via a peptide linker hinge. LPL also contains a large basic patch and a single calcium ion. Additionally, LPL consists of two N-linked [http://en.wikipedia.org/wiki/Glycan glycans] (N70, N386) which likely contribute to the correct folding of LPL due to the attached [http://en.wikipedia.org/wiki/Oligosaccharide oligosaccharides]. Five [http://en.wikipedia.org/wiki/Disulfide disulfide bonds] contribute to the stabilization throughout LPL’s structure. Lastly, the active site in the larger N-terminus domain is lined with hydrophobic residues <ref name="Birrane">PMID:30559189</ref>. | LPL has two main domains, the larger N-terminus domain containing the active site and the smaller C-terminus domain. These two domains are connected via a peptide linker hinge. LPL also contains a large basic patch and a single calcium ion. Additionally, LPL consists of two N-linked [http://en.wikipedia.org/wiki/Glycan glycans] (N70, N386) which likely contribute to the correct folding of LPL due to the attached [http://en.wikipedia.org/wiki/Oligosaccharide oligosaccharides]. Five [http://en.wikipedia.org/wiki/Disulfide disulfide bonds] contribute to the stabilization throughout LPL’s structure. Lastly, the active site in the larger N-terminus domain is lined with hydrophobic residues <ref name="Birrane">PMID:30559189</ref>. | ||
Revision as of 19:10, 27 April 2021
Lipoprotein Lipase (LPL) complexed with GPIHBP1
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Birrane G, Beigneux AP, Dwyer B, Strack-Logue B, Kristensen KK, Francone OL, Fong LG, Mertens HDT, Pan CQ, Ploug M, Young SG, Meiyappan M. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci U S A. 2018 Dec 17. pii: 1817984116. doi:, 10.1073/pnas.1817984116. PMID:30559189 doi:http://dx.doi.org/10.1073/pnas.1817984116
- ↑ 2.0 2.1 Wong H, Davis RC, Thuren T, Goers JW, Nikazy J, Waite M, Schotz MC. Lipoprotein lipase domain function. J Biol Chem. 1994 Apr 8;269(14):10319-23. PMID:8144612
- ↑ Arora R, Nimonkar AV, Baird D, Wang C, Chiu CH, Horton PA, Hanrahan S, Cubbon R, Weldon S, Tschantz WR, Mueller S, Brunner R, Lehr P, Meier P, Ottl J, Voznesensky A, Pandey P, Smith TM, Stojanovic A, Flyer A, Benson TE, Romanowski MJ, Trauger JW. Structure of lipoprotein lipase in complex with GPIHBP1. Proc Natl Acad Sci U S A. 2019 May 21;116(21):10360-10365. doi:, 10.1073/pnas.1820171116. Epub 2019 May 9. PMID:31072929 doi:http://dx.doi.org/10.1073/pnas.1820171116
- ↑ 4.0 4.1 Falko JM. Familial Chylomicronemia Syndrome: A Clinical Guide For Endocrinologists. Endocr Pract. 2018 Aug;24(8):756-763. doi: 10.4158/EP-2018-0157. PMID:30183397 doi:http://dx.doi.org/10.4158/EP-2018-0157
Student/Contributors
- Ashrey Burley
- Allison Welz
- Hannah Wright