We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Betsy Johns/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
===Overview=== | ===Overview=== | ||
| - | DGAT1 is a dimer that has two identical <scene name='87/877512/Labeled_subunits/5'>subunits</scene>. Each subunit contains an MBOAT core that acts as its active site. Each subunit also contains <scene name='87/877512/Labeled_helices/3'>nine transmembrane helices</scene> (TM), 2 intracellular loops (IL), and one ER lumenal loop (EL). <ref name="Wang">PMID: 32433610</ref> TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. A schematic of DGAT1’s structure is shown in Figure 1. | + | DGAT1 is a dimer that has two identical <scene name='87/877512/Labeled_subunits/5'>subunits</scene>. Each subunit contains an MBOAT core that acts as its active site. Each subunit also contains <scene name='87/877512/Labeled_helices/3'>nine transmembrane helices</scene> (TM), 2 intracellular loops (IL), and one ER lumenal loop (EL).<ref name="Wang">PMID: 32433610</ref> TM2-9, IL1, and IL2 form the structure of the MBOAT core active site. A schematic of DGAT1’s structure is shown in Figure 1. |
=== Dimer Interface === | === Dimer Interface === | ||
| Line 23: | Line 23: | ||
===Overview=== | ===Overview=== | ||
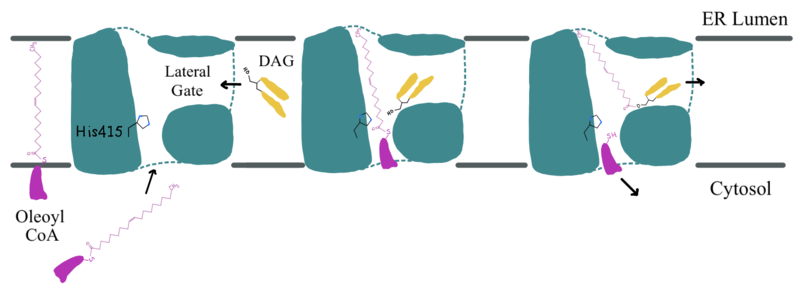
| - | Depiction of the DGAT1 active site mechanism is shown in Figure 2. Oleoyl-CoA, the 18 carbon chain derivative of Acyl-CoA, was used to model the substrate Acyl-CoA. <ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> DAG enters the active site of DGAT1 through | + | Depiction of the DGAT1 active site mechanism is shown in Figure 2. Oleoyl-CoA, the 18 carbon chain derivative of Acyl-CoA, was used to model the substrate Acyl-CoA.<ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> DAG enters the active site of DGAT1 through its lateral gate while the catalytic His415 flips from the cytosolic side to the lumenal side, in order for the channel opening located on the cytosolic side of DGAT1 to widen enough to accommodate the Oleoyl-CoA.<ref name="Sui">PMID: 32433611</ref> The Oleoyl CoA then enters the active site through the channel opening located on the cytosolic side of DGAT1. When the Oleoyl-CoA and DAG are within close proximity in the active site, the catalytic His415 catalyzes the reaction.<ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> The coenzyme A moiety exits through the channel located on the cytosolic side, while the product triacylglyceride exits through the lateral gate. |
| - | [[Image:GDAT_MECH_DRAWING.png|800 px|center|thumb|'''Figure 2: Active Site Mechanism Overview''' Modeled is the active site of DGAT1 (shown in teal) with its catalytic Histidine (His415), Oleoyl CoA (shown in pink), and a general diacylglycerol (DAG, shown in yellow) | + | [[Image:GDAT_MECH_DRAWING.png|800 px|center|thumb|'''Figure 2: Active Site Mechanism Overview''' Modeled is the active site of DGAT1 (shown in teal) with its catalytic Histidine (His415), Oleoyl CoA (shown in pink), and a general diacylglycerol (DAG, shown in yellow).]] |
=== Oleoyl-CoA Binding === | === Oleoyl-CoA Binding === | ||
| - | Oleoyl-CoA enters DGAT1’s active site through a channel on the cytosolic side of the membrane. In order for the channel to widen and accommodate the fatty acid tail of the Oleoyl-CoA, <scene name='87/877512/Dgat_aligned2/17'>His415</scene> must flip from facing the cytosolic side to facing the lumenal side. It is speculated that the delta sulfur of the Met434 is involved in hydrogen bonding interactions with the catalytic His415 while it is flipped toward the cytosolic side of DGAT1 ([https://www.rcsb.org/structure/6VYI 6vyi]). <ref name="Sui">PMID: 32433611</ref> The His415 needs to break these speculated hydrogen bonds with Met434 to flip toward the lumenal side of DGAT1. Once <scene name='87/877512/Acyl_coa/2'>Oleoyl-CoA</scene> is bound in the active site, residues Asn378, Gln437, Met434, and Gln465 stabilize the fatty acid tail within the cytosolic channel of the active site, while residues His415 and Gln416 are directly involved within the catalytic mechanism of DGAT1. <ref name="Sui">PMID: 32433611</ref> | + | Oleoyl-CoA enters DGAT1’s active site through a channel on the cytosolic side of the membrane. In order for the channel to widen and accommodate the fatty acid tail of the Oleoyl-CoA, <scene name='87/877512/Dgat_aligned2/17'>His415</scene> must flip from facing the cytosolic side to facing the lumenal side. It is speculated that the delta sulfur of the Met434 is involved in hydrogen bonding interactions with the catalytic His415 while it is flipped toward the cytosolic side of DGAT1 ([https://www.rcsb.org/structure/6VYI 6vyi]).<ref name="Sui">PMID: 32433611</ref> The His415 needs to break these speculated hydrogen bonds with Met434 to flip toward the lumenal side of DGAT1. Once <scene name='87/877512/Acyl_coa/2'>Oleoyl-CoA</scene> is bound in the active site, residues Asn378, Gln437, Met434, and Gln465 stabilize the fatty acid tail within the cytosolic channel of the active site, while residues His415 and Gln416 are directly involved within the catalytic mechanism of DGAT1.<ref name="Sui">PMID: 32433611</ref> |
=== DAG Binding === | === DAG Binding === | ||
| - | <scene name='87/877512/Dag_binding/2'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter. <ref name="Sui">PMID: 32433611</ref> The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with <scene name='87/877512/Lateral_gate_residues/4'>hydrophobic residues</scene> Phe342, Leu261, and Val381. <ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. | + | <scene name='87/877512/Dag_binding/2'>DAG</scene> enters the active site through the lateral gate located in the lipid bilayer of the membrane. This lateral gate is a bent and hydrophobic channel that allows for hydrophobic linear or curvilinear molecules to enter.<ref name="Sui">PMID: 32433611</ref> The lateral gate channel is designed to allow for the entrance of DAG and the exit of a triacylglyceride. This channel is also lined with <scene name='87/877512/Lateral_gate_residues/4'>hydrophobic residues</scene> Phe342, Leu261, and Val381.<ref name="Wang">PMID: 32433610</ref><ref name="Sui">PMID: 32433611</ref> Once within the channel, DAG is positioned in close proximity to the bound Acyl-CoA and the catalytic His415. |
=== Active Site Structure === | === Active Site Structure === | ||
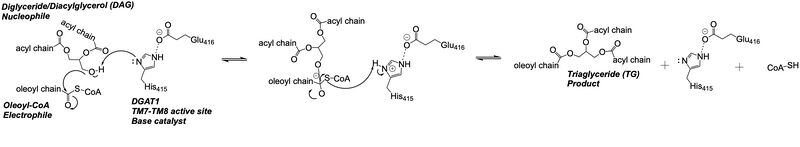
| - | The <scene name='87/877512/Active_site_zoomed_out/4'>active site</scene> is located within the pocket formed from TM2-TM9, IL1, and IL2, with the catalytic residue Histidine 415 located on TM7. It is accessible through openings both on the cytosolic and luminal sides. The active site of DGAT1 serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and bind the fatty acid to the diacylglycerol. The conserved His415 acts catalytically by a [https://www.genscript.com/molecular-biology-glossary/478/charge-relay-system#:~:text=A%20tautomeric%20form%20of%20a,another%20in%20the%20same%20protein charge relay system], where the negative charge of the neighboring Glu416 pulls on the electrons of histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through [https://chem.libretexts.org/Bookshelves/Organic_Chemistry/Map%3A_Organic_Chemistry_(McMurry)/21%3A_Carboxylic_Acid_Derivatives-_Nucleophilic_Acyl_Substitution_Reactions/21.02%3A_Nucleophilic_Acyl_Substitution_Reactions acyl substitution]. The catalytic mechanism for DGAT1 is shown in Figure 3. <ref name="Wang">PMID: 32433610</ref> | + | The <scene name='87/877512/Active_site_zoomed_out/4'>active site</scene> is located within the pocket formed from TM2-TM9, IL1, and IL2, with the catalytic residue Histidine 415 located on TM7. It is accessible through openings both on the cytosolic and luminal sides. The active site of DGAT1 serves its catalytic function by placing the His415 residue in close proximity to the acyl-CoA in order to cleave its ester bond and bind the fatty acid to the diacylglycerol. The conserved His415 acts catalytically by a [https://www.genscript.com/molecular-biology-glossary/478/charge-relay-system#:~:text=A%20tautomeric%20form%20of%20a,another%20in%20the%20same%20protein charge relay system], where the negative charge of the neighboring Glu416 pulls on the electrons of histidine at the N1 position, making the N3 position more nucleophilic. This nitrogen will then deprotonate DAG so it can begin its attack on Acyl-CoA through [https://chem.libretexts.org/Bookshelves/Organic_Chemistry/Map%3A_Organic_Chemistry_(McMurry)/21%3A_Carboxylic_Acid_Derivatives-_Nucleophilic_Acyl_Substitution_Reactions/21.02%3A_Nucleophilic_Acyl_Substitution_Reactions acyl substitution]. The catalytic mechanism for DGAT1 is shown in Figure 3.<ref name="Wang">PMID: 32433610</ref> |
The oxyanion hole of the acyl substitution intermediate should be stabilized by DGAT1. However, the two available PDB files for the substrate-bound DGAT1, [https://www.rcsb.org/structure/6VZ1 6vz1] and [https://www.rcsb.org/structure/6VP0 6vp0], do not show a possible stabilizing residue for the Oleoyl-CoA, the 18 carbon chain version of Acyl-CoA. The PDB file 6vz1 shows the sulfur of Oleoyl-CoA, the electrophile of the mechanism, <scene name='87/877512/Active_site_zoomed_in_6vz1/3'>facing away from His415</scene>. This is not likely to be the proper orientation for Oleoyl-CoA, as this sulfur would have to be facing towards the His415 to be protonated after the acyl substitution. For an oxyanion stabilizing residue to be located within this PDB file, the Oleoyl-CoA would have to be properly oriented. The other substrate-bound PDB file 6vp0 shows the sulfur of Oleoyl-CoA facing toward the His415. However, <scene name='87/877512/Active_site_zoomed_in_6vp0/2'>the tail of oleoyl-CoA is bent</scene> and curved around the oxyanion hole, preventing a stabilizing residue within DGAT1 to be found. For these reasons, an oxyanion stabilizing residue was not identified in the mechanism nor in the images. | The oxyanion hole of the acyl substitution intermediate should be stabilized by DGAT1. However, the two available PDB files for the substrate-bound DGAT1, [https://www.rcsb.org/structure/6VZ1 6vz1] and [https://www.rcsb.org/structure/6VP0 6vp0], do not show a possible stabilizing residue for the Oleoyl-CoA, the 18 carbon chain version of Acyl-CoA. The PDB file 6vz1 shows the sulfur of Oleoyl-CoA, the electrophile of the mechanism, <scene name='87/877512/Active_site_zoomed_in_6vz1/3'>facing away from His415</scene>. This is not likely to be the proper orientation for Oleoyl-CoA, as this sulfur would have to be facing towards the His415 to be protonated after the acyl substitution. For an oxyanion stabilizing residue to be located within this PDB file, the Oleoyl-CoA would have to be properly oriented. The other substrate-bound PDB file 6vp0 shows the sulfur of Oleoyl-CoA facing toward the His415. However, <scene name='87/877512/Active_site_zoomed_in_6vp0/2'>the tail of oleoyl-CoA is bent</scene> and curved around the oxyanion hole, preventing a stabilizing residue within DGAT1 to be found. For these reasons, an oxyanion stabilizing residue was not identified in the mechanism nor in the images. | ||
| Line 50: | Line 50: | ||
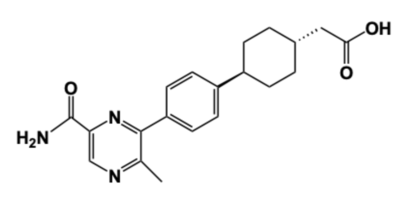
[[Image:DGAT_INHIBITOR.png|400 px|right|thumb|'''Figure 4: DGAT1 Inhibitor AZD7687''' Shown is the structure of AZD7687, a known inhibitor of DGAT1.]] | [[Image:DGAT_INHIBITOR.png|400 px|right|thumb|'''Figure 4: DGAT1 Inhibitor AZD7687''' Shown is the structure of AZD7687, a known inhibitor of DGAT1.]] | ||
| - | Obesity and nonalcoholic fatty liver disease ([https://www.mayoclinic.org/diseases-conditions/nonalcoholic-fatty-liver-disease/symptoms-causes/syc-20354567 NAFLD]) result from an accumulation of triacylglycerides within the body. Recently, DGAT1 has become a therapeutic target for obesity and nonalcoholic fatty liver disease in order to reduce triacylglyceride storage within the body. <ref name="Denison">PMID: 24118885</ref><ref name="Villanueva">PMID: 19472314</ref> Different inhibitors have been created, such as AstraZeneca’s direct inhibitor [https://www.apexbt.com/azd7687.html AZD7687], shown in Figure 4. <ref name="Denison">PMID: 24118885</ref> AZD7687 has an EC50 value 0.44 µmol/L, showing that it binds with high affinity at a low concentration of DGAT1. <ref name="Denison">PMID: 24118885</ref> However, while triacylglyceride accumulation decreased, negative side effects did occur, such as diarrhea and other adverse GI symptoms. <ref name="Denison">PMID: 24118885</ref> | + | Obesity and nonalcoholic fatty liver disease ([https://www.mayoclinic.org/diseases-conditions/nonalcoholic-fatty-liver-disease/symptoms-causes/syc-20354567 NAFLD]) result from an accumulation of triacylglycerides within the body. Recently, DGAT1 has become a therapeutic target for obesity and nonalcoholic fatty liver disease in order to reduce triacylglyceride storage within the body.<ref name="Denison">PMID: 24118885</ref><ref name="Villanueva">PMID: 19472314</ref> Different inhibitors have been created, such as AstraZeneca’s direct inhibitor [https://www.apexbt.com/azd7687.html AZD7687], shown in Figure 4.<ref name="Denison">PMID: 24118885</ref> AZD7687 has an EC50 value 0.44 µmol/L, showing that it binds with high affinity at a low concentration of DGAT1.<ref name="Denison">PMID: 24118885</ref> However, while triacylglyceride accumulation decreased, negative side effects did occur, such as diarrhea and other adverse GI symptoms.<ref name="Denison">PMID: 24118885</ref> |
===Congenital Protein-Losing Enteropathy=== | ===Congenital Protein-Losing Enteropathy=== | ||
| - | Additionally, congenital protein-losing enteropathy ([https://www.uptodate.com/contents/protein-losing-gastroenteropathy PLE]) is linked to DGAT1 mutations. PLE is a GI disorder that causes malabsorption of fat and a deficiency in fat-soluble vitamins. Patients in a congenital PLE case study exhibited a homozygous missense Leu295Pro mutation within the MBOAT core of their DGAT1 enzymes. <ref name="Stephen">PMID: 26883093</ref> <scene name='87/877512/Mutation/6'>Leu295</scene> is located within the MBOAT core active site on TM5. While the Leu295 is not near the catalytic residues His415 and Glu416, the <scene name='88/880292/Cple/6'>Leu295Pro</scene> mutation will disrupt the overall active site. Proline is an alpha helix breaker because it causes steric hindrance within the backbone of the helix turn. It is hypothesized that this mutation breaks this helix in the MBOAT core and greatly reduces its enzymatic activity and ability to make triacylglycerides. Without proper DGAT1 function to produce triacylglycerides, there is a decrease in albumin, which is a protein that helps prevent fluid from leaking out of the liver and blood vessels. <ref name="Villanueva">PMID: 19472314</ref> This decrease in [https://medlineplus.gov/lab-tests/albumin-blood-test/#:~:text=Albumin%20is%20a%20protein%20made,with%20your%20liver%20or%20kidneys albumin] then leads to decreased efficiency in nutrient transport and fat absorption. | + | Additionally, congenital protein-losing enteropathy ([https://www.uptodate.com/contents/protein-losing-gastroenteropathy PLE]) is linked to DGAT1 mutations. PLE is a GI disorder that causes malabsorption of fat and a deficiency in fat-soluble vitamins. Patients in a congenital PLE case study exhibited a homozygous missense Leu295Pro mutation within the MBOAT core of their DGAT1 enzymes.<ref name="Stephen">PMID: 26883093</ref> <scene name='87/877512/Mutation/6'>Leu295</scene> is located within the MBOAT core active site on TM5. While the Leu295 is not near the catalytic residues His415 and Glu416, the <scene name='88/880292/Cple/6'>Leu295Pro</scene> mutation will disrupt the overall active site. Proline is an alpha helix breaker because it causes steric hindrance within the backbone of the helix turn. It is hypothesized that this mutation breaks this helix in the MBOAT core and greatly reduces its enzymatic activity and ability to make triacylglycerides. Without proper DGAT1 function to produce triacylglycerides, there is a decrease in albumin, which is a protein that helps prevent fluid from leaking out of the liver and blood vessels.<ref name="Villanueva">PMID: 19472314</ref> This decrease in [https://medlineplus.gov/lab-tests/albumin-blood-test/#:~:text=Albumin%20is%20a%20protein%20made,with%20your%20liver%20or%20kidneys albumin] then leads to decreased efficiency in nutrient transport and fat absorption. |
===Relevance=== | ===Relevance=== | ||
Revision as of 20:47, 27 April 2021
Diacylglycerol acyltransferase 1, DGAT1, synthesizes triacylglycerides
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Wang L, Qian H, Nian Y, Han Y, Ren Z, Zhang H, Hu L, Prasad BVV, Laganowsky A, Yan N, Zhou M. Structure and mechanism of human diacylglycerol O-acyltransferase 1. Nature. 2020 May;581(7808):329-332. doi: 10.1038/s41586-020-2280-2. Epub 2020 May, 13. PMID:32433610 doi:http://dx.doi.org/10.1038/s41586-020-2280-2
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 Sui X, Wang K, Gluchowski NL, Elliott SD, Liao M, Walther TC, Farese RV Jr. Structure and catalytic mechanism of a human triacylglycerol-synthesis enzyme. Nature. 2020 May;581(7808):323-328. doi: 10.1038/s41586-020-2289-6. Epub 2020 May, 13. PMID:32433611 doi:http://dx.doi.org/10.1038/s41586-020-2289-6
- ↑ 3.0 3.1 3.2 Ma D, Wang Z, Merrikh CN, Lang KS, Lu P, Li X, Merrikh H, Rao Z, Xu W. Crystal structure of a membrane-bound O-acyltransferase. Nature. 2018 Oct;562(7726):286-290. doi: 10.1038/s41586-018-0568-2. Epub 2018 Oct, 3. PMID:30283133 doi:http://dx.doi.org/10.1038/s41586-018-0568-2
- ↑ 4.0 4.1 4.2 4.3 Denison H, Nilsson C, Lofgren L, Himmelmann A, Martensson G, Knutsson M, Al-Shurbaji A, Tornqvist H, Eriksson JW. Diacylglycerol acyltransferase 1 inhibition with AZD7687 alters lipid handling and hormone secretion in the gut with intolerable side effects: a randomized clinical trial. Diabetes Obes Metab. 2014 Apr;16(4):334-43. doi: 10.1111/dom.12221. Epub 2013 Oct, 31. PMID:24118885 doi:http://dx.doi.org/10.1111/dom.12221
- ↑ 5.0 5.1 Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, Farese RV Jr. Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology. 2009 Aug;50(2):434-42. doi: 10.1002/hep.22980. PMID:19472314 doi:http://dx.doi.org/10.1002/hep.22980
- ↑ Stephen J, Vilboux T, Haberman Y, Pri-Chen H, Pode-Shakked B, Mazaheri S, Marek-Yagel D, Barel O, Di Segni A, Eyal E, Hout-Siloni G, Lahad A, Shalem T, Rechavi G, Malicdan MC, Weiss B, Gahl WA, Anikster Y. Congenital protein losing enteropathy: an inborn error of lipid metabolism due to DGAT1 mutations. Eur J Hum Genet. 2016 Aug;24(9):1268-73. doi: 10.1038/ejhg.2016.5. Epub 2016 Feb , 17. PMID:26883093 doi:http://dx.doi.org/10.1038/ejhg.2016.5
Student Contributors
- Betsy Johns
- Elise Wang
- Tyler Bihasa