Sandbox GGC3
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
ANL enzymes follow a domain alternation strategy for the first adenylation reaction, in which the reaction is catalyzed by <scene name='75/752266/4g36/2'>one conformation</scene> and following the formation of the adenylate intermediate and release of pyrophosphate (PPi), the C-terminal domain undergoes a rotational transformation that is necessary for <scene name='75/752266/4g37/2'>the second partial reaction</scene>. The <scene name='75/752266/Active_site/1'>active site</scene><ref name=“Branchini”>Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. ''Biochemistry 38''(40), 13223–13230. https://doi.org/10.1021/bi991181o</ref> of ANL enzymes resides between a 400-500 residue N-terminal domain and a smaller C-terminal domain of ~110-130 amino acids<ref name="Sundlov">Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. ''Biochemistry 51''(33), 6493-6495. https://doi.org/10.1021/bi300934s</ref>. Ten conserved regions of these proteins have been termed the A1-A10 motifs which play critical roles in either or both partial reactions<ref name="Marahiel">Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. ''Chemical Reviews 97''(7), 2651-2674. https://doi.org/10.1021/cr960029e</ref>. Two lysine residues are required for each partial reaction, suggestive that luciferase similarly adopts a rotational transformation for complete catalysis. A mutation of <scene name='75/752266/Lys529/1'>Lys529</scene>, the A10 lysine, impairs only the initial adenylation reaction<ref name="Sundlov"/> whereas mutation of <scene name='75/752266/Lys443/1'>Lys443</scene> in the A8 region disrupts the oxidative reaction<ref name="Sundlov"/>. | ANL enzymes follow a domain alternation strategy for the first adenylation reaction, in which the reaction is catalyzed by <scene name='75/752266/4g36/2'>one conformation</scene> and following the formation of the adenylate intermediate and release of pyrophosphate (PPi), the C-terminal domain undergoes a rotational transformation that is necessary for <scene name='75/752266/4g37/2'>the second partial reaction</scene>. The <scene name='75/752266/Active_site/1'>active site</scene><ref name=“Branchini”>Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. ''Biochemistry 38''(40), 13223–13230. https://doi.org/10.1021/bi991181o</ref> of ANL enzymes resides between a 400-500 residue N-terminal domain and a smaller C-terminal domain of ~110-130 amino acids<ref name="Sundlov">Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. ''Biochemistry 51''(33), 6493-6495. https://doi.org/10.1021/bi300934s</ref>. Ten conserved regions of these proteins have been termed the A1-A10 motifs which play critical roles in either or both partial reactions<ref name="Marahiel">Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. ''Chemical Reviews 97''(7), 2651-2674. https://doi.org/10.1021/cr960029e</ref>. Two lysine residues are required for each partial reaction, suggestive that luciferase similarly adopts a rotational transformation for complete catalysis. A mutation of <scene name='75/752266/Lys529/1'>Lys529</scene>, the A10 lysine, impairs only the initial adenylation reaction<ref name="Sundlov"/> whereas mutation of <scene name='75/752266/Lys443/1'>Lys443</scene> in the A8 region disrupts the oxidative reaction<ref name="Sundlov"/>. | ||
| - | |||
=== Biochemical Mechanism of LH2-AMP Oxidation=== | === Biochemical Mechanism of LH2-AMP Oxidation=== | ||
| Line 40: | Line 39: | ||
== Structural highlights == | == Structural highlights == | ||
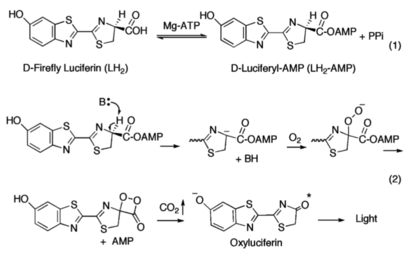
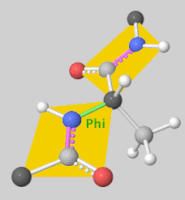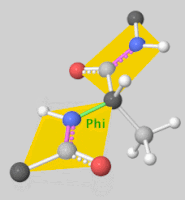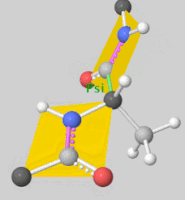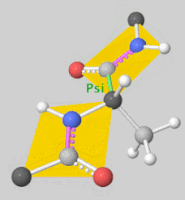
| + | [[Image:DLSA.png|thumb|right|upright=2.3|5'-O-[N-(Dehydroluciferyl)-sulfamoyl] adenosine, shortened to DLSA for brevity. The sulfamate moiety is shown to the left (sulfur atoms are represented in yellow while oxygen atoms are represented in yellow. Further, the carbonyl oxygen of the luciferyl-adenylate is connected to the sulfamate moiety via nitrogen atom (represented in blue). [https://pubchem.ncbi.nlm.nih.gov/compound/5_-O-_N-_Dehydroluciferyl_-sulfamoyl_-adenosine]]][[Image:Dihedral-angles-anim.gif|thumb|left|upright=5.1|The φ(Phi)/ψ(Psi) angles of the Lys439 residue undergo a rotational transformation of −73°/−12° to −69°/158° in the adenylate-forming to second-catalytic conformation, respectively, showing that the major torsional change in the ψ angle is observed<ref name="Sundlov"/>.]] | ||
| - | [[Image:Dihedral-angles-anim.gif|thumb|right|upright=5.1|The φ(Phi)/ψ(Psi) angles of the Lys439 residue undergo a rotational transformation of −73°/−12° to −69°/158° in the adenylate-forming to second-catalytic conformation, respectively, showing that the major torsional change in the ψ angle is observed<ref name="Sundlov"/>.]] | ||
The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, <scene name='75/752266/Nlys529oogly316/1'>interacts</scene> with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the <scene name='75/752266/N_lys443/1'>side chain amine</scene> of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it <scene name='75/752266/Gln448ando/1'>interacts</scene> with a sulfamate oxygen<ref name="Sundlov"/><ref name="Bruce"/>. Altogether (with the inclusion of an <scene name='75/752266/Glu479andarg437/1'>ionic interaction</scene> between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. | The conserved catalytic lysine for the adenylation reaction<ref name="conserved">Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. ''Biochemistry 39''(18), 5433-5440. https://doi.org/10.1021/bi9928804</ref>, Lys529, <scene name='75/752266/Nlys529oogly316/1'>interacts</scene> with the carbonyl oxygen of the adenylate, the O5 atom that bridges the ribose and sulfamate moiety, and the main chain carbonyl of Gly316. The second conformation observations show that the <scene name='75/752266/N_lys443/1'>side chain amine</scene> of Lys443 adopts a nearly identical position as Lys529, and Gln448 of the C-terminal domain rotates into the binding pocket where it <scene name='75/752266/Gln448ando/1'>interacts</scene> with a sulfamate oxygen<ref name="Sundlov"/><ref name="Bruce"/>. Altogether (with the inclusion of an <scene name='75/752266/Glu479andarg437/1'>ionic interaction</scene> between Glu479 and Arg437), these interactions are responsible for the stabilization of the new C-terminal conformation. | ||
| + | |||
== Relevance == | == Relevance == | ||
Revision as of 22:12, 27 April 2021
Firefly Luciferase
apple juice! B~)
| |||||||||||
References
- ↑ Branchini, B. R., Magyar, R. A., Murtiashaw, M. H., Anderson, S. M., Helgerson, L. C., & Zimmer, M. (1999). Site-directed mutagenesis of firefly luciferase active site amino acids: a proposed model for bioluminescence color. Biochemistry 38(40), 13223–13230. https://doi.org/10.1021/bi991181o
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Sundlov, J. A., Fontaine, D. M., Southworth, T. L., Branchini, B. R., Gulick, A. M. (2012). Crystal Structure of Firefly Luciferase in a Second Catalytic Conformation Supports a Domain Alternation Mechanism. Biochemistry 51(33), 6493-6495. https://doi.org/10.1021/bi300934s
- ↑ Marahiel, M. A., Stachelhaus, T., Mootz, H. D. (1997). Modular Peptide Synthetases Involved in Nonribosmal Peptide Synthesis. Chemical Reviews 97(7), 2651-2674. https://doi.org/10.1021/cr960029e
- ↑ 4.0 4.1 Branchini, B. R., Southworth, T. L., Murtiahsaw, M. H., Wilkinson, S. R., Khattak, N. F., Rosenberg, J. C., & Zimmer, M. (2005). Mutagenesis Evidence that the Partial Reactions of Firefly Bioluminescence are Catalyzed by Different Conformations of the Luciferase C-Terminal Domain. “Biochemistry 44”(5), 1385-1393. https://doi.org/10.1021/bi047903f
- ↑ Oba, Y., Ojika, M., Inouye, S. (2003). Firefly luciferase is a bifunctional enzyme: ATP-dependent monoxygenase and a long chain fatty acyl-CoA synthetase. “FEBS Letters 540”(1-3), 251-254. https://doi.org/10.1016/S0014-5793(03)00272-2
- ↑ Nakamura, M., Maki, S., Amano, Y., Ohkita, Y., Niwa, K., Hirano, T., Ohmiya, Y., & Niwa, H. (2005). Firefly luciferase exhibits bimodal action depending on the luciferin chirality. “Biochemical and Biophysical Research Communications, 331”(2), 471–475. https://doi.org/10.1016/j.bbrc.2005.03.202
- ↑ Branchini, B. R., Murtiashaw, M. H., Magyar, R. A., Anderson, S. M. (2000). The Role of Lysine 529, a Conserved Residue of the Acyl-Adenylate-Forming Enzyme Superfamily, in Firefly Luciferase. Biochemistry 39(18), 5433-5440. https://doi.org/10.1021/bi9928804
- ↑ Sala-Newby, G. B., & Campbell, A. K. (1991). Engineering a bioluminescent indicator for cyclic AMP-dependent protein kinase. “The Biochemical Journal”, 279 (Pt 3), 727–732. https://doi.org/10.1042/bj2790727
- ↑ de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., & Subramani, S. (1987). Firefly luciferase gene: structure and expression in mammalian cells. Molecular and cellular biology, 7(2), 725–737. https://doi.org/10.1128/mcb.7.2.725
- ↑ de Wet, J. R., Wood, K. V., Helinski, D. R., & DeLuca, M. (1985). Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America, 82(23), 7870–7873. https://doi.org/10.1073/pnas.82.23.7870
- ↑ Thorne, N., Shen, M., Lea, W. A., Simeonov, A., Lovell, S., Auld, D. S., & Inglese, J. (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chemistry & biology, 19(8), 1060–1072. https://doi.org/10.1016/j.chembiol.2012.07.015
![The Common Eastern Firefly in a hand emitting a yellow hue, showing bioluminescence.[1]](/wiki/images/thumb/6/62/Common_Eastern_Firefly.jpg/200px-Common_Eastern_Firefly.jpg)
![5'-O-[N-(Dehydroluciferyl)-sulfamoyl] adenosine, shortened to DLSA for brevity. The sulfamate moiety is shown to the left (sulfur atoms are represented in yellow while oxygen atoms are represented in yellow. Further, the carbonyl oxygen of the luciferyl-adenylate is connected to the sulfamate moiety via nitrogen atom (represented in blue). [2]](/wiki/images/thumb/6/61/DLSA.png/410px-DLSA.png)
![The Common Eastern Firefly expressing bioluminescence seen giving off a yellow-green hue.[3]](/wiki/images/thumb/e/e9/CommonEasternFirefly.jpg/320px-CommonEasternFirefly.jpg)
