1a0a
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1a0a' size='340' side='right'caption='[[1a0a]], [[Resolution|resolution]] 2.80Å' scene=''> | <StructureSection load='1a0a' size='340' side='right'caption='[[1a0a]], [[Resolution|resolution]] 2.80Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1a0a]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1a0a]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1A0A OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1A0A FirstGlance]. <br> |
| - | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1a0a FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1a0a OCA], [https://pdbe.org/1a0a PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1a0a RCSB], [https://www.ebi.ac.uk/pdbsum/1a0a PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1a0a ProSAT]</span></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.8Å</td></tr> |
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1a0a FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1a0a OCA], [https://pdbe.org/1a0a PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1a0a RCSB], [https://www.ebi.ac.uk/pdbsum/1a0a PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1a0a ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
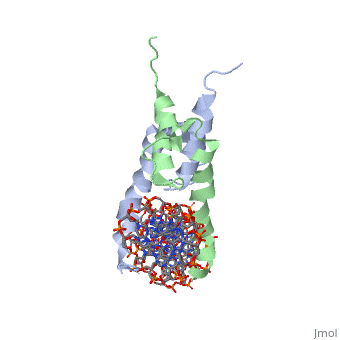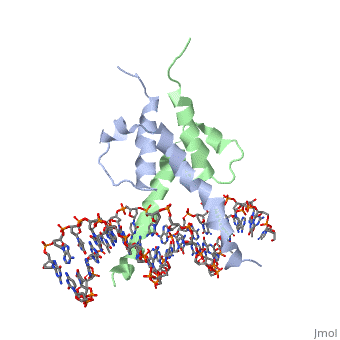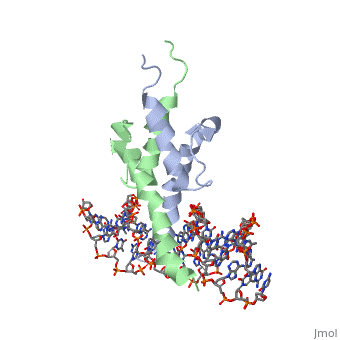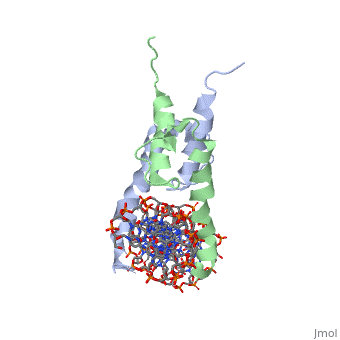
| - | + | [https://www.uniprot.org/uniprot/PHO4_YEAST PHO4_YEAST] Transcriptional activator that regulates the expression of repressible phosphatase under phosphate starvation conditions. Binds to the upstream activating sequence (UAS) of several phosphatase encoding PHO genes. Inhibited by the cyclin-CDK PHO80-PHO85 under high-phosphate conditions. | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 18: | Line 19: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1a0a ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1a0a ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The crystal structure of a DNA-binding domain of PHO4 complexed with DNA at 2.8 A resolution revealed that the domain folds into a basic-helix-loop-helix (bHLH) motif with a long but compact loop that contains a short alpha-helical segment. This helical structure positions a tryptophan residue into an aromatic cluster so as to make the loop compact. PHO4 binds to DNA as a homodimer with direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The 3'-flanking bases GG are recognized by Arg2 and His5. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein. | ||
| - | |||
| - | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.,Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313<ref>PMID:9303313</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1a0a" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Pho4|Pho4]] | *[[Pho4|Pho4]] | ||
| - | == References == | ||
| - | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: Atcc 18824]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Hakoshima | + | [[Category: Saccharomyces cerevisiae]] |
| - | [[Category: Ihara | + | [[Category: Hakoshima T]] |
| - | [[Category: Kyogoku | + | [[Category: Ihara K]] |
| - | [[Category: Ogawa | + | [[Category: Kyogoku Y]] |
| - | [[Category: Oshima | + | [[Category: Ogawa N]] |
| - | [[Category: Shimizu | + | [[Category: Oshima Y]] |
| - | [[Category: Shimizu | + | [[Category: Shimizu M]] |
| - | [[Category: Toumoto | + | [[Category: Shimizu T]] |
| - | + | [[Category: Toumoto A]] | |
| - | + | ||
| - | + | ||
Current revision
PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX
| |||||||||||