Histone deacetylase 8 (HDAC8)
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
== Introduction == | == Introduction == | ||
Histone deacetylase 8 (<scene name=83/834033/Overall_structure_new/1'>HDAC8</scene>) is an enzyme that plays a role in controlling gene expression. Specifically, HDAC8 removes an acetyl group from the ε-amino-Lys 382 of the N-terminal extension of Histone 4. <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> [https://proteopedia.org/wiki/index.php/Nucleosome Histone] core particles consist of eight monomer proteins that an octamer complex. Each histone monomer has a net positive charge which enhances the interaction with negatively-charged DNA. This prevents transcription factors from accessing DNA, thus decreasing gene expression. [https://en.wikipedia.org/wiki/Chromatin_remodeling Chromatin remodeling] by the addition or removal of an acetyl group on a histone is an example of [https://en.wikipedia.org/wiki/Epigenetics epigenetic regulation]. [https://en.wikipedia.org/wiki/Histone_acetyltransferase Histone Acetylase 1] (HAT1) catalyzes the addition of an acetyl group onto a lysine residue in a histone. The lack of charge on the acetylated lysine weakens the interaction between DNA and histones. This subsequently allows transcription factors to access to bind DNA, increasing the potential for gene expression. HDAC8 reverses this reaction by catalyzing the removal of these acetyl groups from the Lys to reclaim the positive charge of the histone. This again allows the histone to interact with the negative charge on the DNA. As a result, DNA binds more tightly to the histone protein, repressing transcription. | Histone deacetylase 8 (<scene name=83/834033/Overall_structure_new/1'>HDAC8</scene>) is an enzyme that plays a role in controlling gene expression. Specifically, HDAC8 removes an acetyl group from the ε-amino-Lys 382 of the N-terminal extension of Histone 4. <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> [https://proteopedia.org/wiki/index.php/Nucleosome Histone] core particles consist of eight monomer proteins that an octamer complex. Each histone monomer has a net positive charge which enhances the interaction with negatively-charged DNA. This prevents transcription factors from accessing DNA, thus decreasing gene expression. [https://en.wikipedia.org/wiki/Chromatin_remodeling Chromatin remodeling] by the addition or removal of an acetyl group on a histone is an example of [https://en.wikipedia.org/wiki/Epigenetics epigenetic regulation]. [https://en.wikipedia.org/wiki/Histone_acetyltransferase Histone Acetylase 1] (HAT1) catalyzes the addition of an acetyl group onto a lysine residue in a histone. The lack of charge on the acetylated lysine weakens the interaction between DNA and histones. This subsequently allows transcription factors to access to bind DNA, increasing the potential for gene expression. HDAC8 reverses this reaction by catalyzing the removal of these acetyl groups from the Lys to reclaim the positive charge of the histone. This again allows the histone to interact with the negative charge on the DNA. As a result, DNA binds more tightly to the histone protein, repressing transcription. | ||
| - | |||
| - | See [[Histone deacetylase]]. | ||
==HDAC Enzymes and Homology== | ==HDAC Enzymes and Homology== | ||
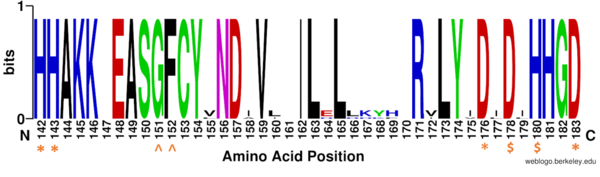
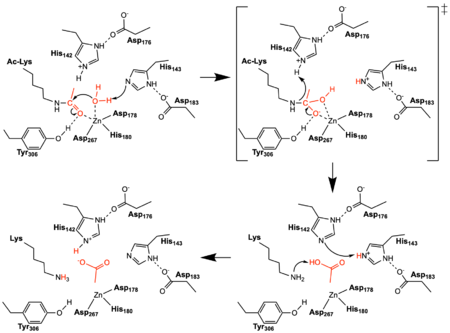
| - | There are four major classes of HDAC proteins (I,II, III, and IV). Other than the Class III “[https://en.wikipedia.org/wiki/Sirtuin sirtuins]” that utilize a NAD<sup>+</sup> cofactor-dependent mechanism <ref name="Sauve">PMID:23102634</ref>, all other HDAC classes use Zn<sup>2+</sup>-assisted catalysis to activate a water molecule for nucleophilic attack (Figure 2).<ref name="DesJarlais, R., & Tummino, P. J.">DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210 </ref> While Classes I, II, and IV do have some major distinctions such as size of the protein, in general, they share homology at the catalytic site. HDAC8 is classified as a Class I HDAC alongside HDACs 1-3. In fact, within Class I HDACs, there are many invariant residues involved in the catalytic site (such as His-Asp dyads), Zn-binding, and ligand binding pocket (such as Asp101) (Figure 1). <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> The structure reported here is of Human HDAC8, a 377 residue, Class I HDAC. | + | There are four major classes of [https://proteopedia.org/wiki/index.php/Histone_deacetylase HDAC] proteins (I,II, III, and IV). Other than the Class III “[https://en.wikipedia.org/wiki/Sirtuin sirtuins]” that utilize a NAD<sup>+</sup> cofactor-dependent mechanism <ref name="Sauve">PMID:23102634</ref>, all other HDAC classes use Zn<sup>2+</sup>-assisted catalysis to activate a water molecule for nucleophilic attack (Figure 2).<ref name="DesJarlais, R., & Tummino, P. J.">DesJarlais, R., & Tummino, P. J. (2016). Role of histone-modifying enzymes and their complexes in regulation of chromatin biology. Biochemistry, 55(11), 1584-1599. https://doi.org/10.1021/acs.biochem.5b01210 </ref> While Classes I, II, and IV do have some major distinctions such as size of the protein, in general, they share homology at the catalytic site. HDAC8 is classified as a Class I HDAC alongside HDACs 1-3. In fact, within Class I HDACs, there are many invariant residues involved in the catalytic site (such as His-Asp dyads), Zn-binding, and ligand binding pocket (such as Asp101) (Figure 1). <ref name="Vannini, A., Volpari, C., Gallinari, P.">Vannini, A., Volpari, C., Gallinari, P., Jones, P., Mattu, M., Carfí, A., ... & Di Marco, S. (2007). Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8–substrate complex. EMBO reports, 8(9), 879-884. https://doi.org/10.1038/sj.embor.7401047 </ref> The structure reported here is of Human HDAC8, a 377 residue, Class I HDAC. |
[[Image:Conserved residues.PNG|600px||right||thumb|Figure 1: Weblogo representation comparing conservation of residues (143-182 in HDAC8) to homologous sequences in all class I HDACs. Active site residues (asterisk), zinc binding (dollar), and binding pocket residues (caret) are invariant across class I HDACs. Other conserved residues not shown include active site residue Tyr306, zinc binding residue Asp267, and binding pocket residue Asp101. Other conserved residues in the Weblogo image serve as a 'scaffold' for the active site residues, while <scene name='83/834033/Non-conserved_helix/2'>non-conserved</scene> residues from 158 to 170 are part of an α-helix that transits away from the active site before looping back to it.]] | [[Image:Conserved residues.PNG|600px||right||thumb|Figure 1: Weblogo representation comparing conservation of residues (143-182 in HDAC8) to homologous sequences in all class I HDACs. Active site residues (asterisk), zinc binding (dollar), and binding pocket residues (caret) are invariant across class I HDACs. Other conserved residues not shown include active site residue Tyr306, zinc binding residue Asp267, and binding pocket residue Asp101. Other conserved residues in the Weblogo image serve as a 'scaffold' for the active site residues, while <scene name='83/834033/Non-conserved_helix/2'>non-conserved</scene> residues from 158 to 170 are part of an α-helix that transits away from the active site before looping back to it.]] | ||
Revision as of 14:44, 14 June 2021
Histone Deacetylase 8: A zinc-dependent hydrolase and transcriptional silencer
| |||||||||||
Student Contributors
Courtney Brown, Asif Hossain, Carolyn Hurdle, Cassandra Marsh, Sean O'Brien, and Josephine Thestrup
Proteopedia Page Contributors and Editors (what is this?)
Mark Macbeth, Angel Herraez, Valentine J Klimkowski, Michal Harel