We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1ls8
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
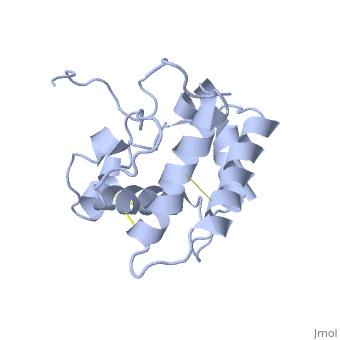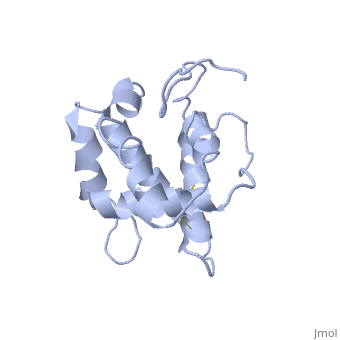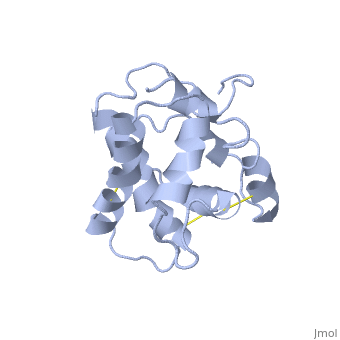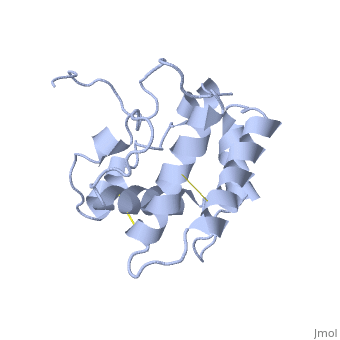
==NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH== | ==NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH== | ||
| - | <StructureSection load='1ls8' size='340' side='right'caption='[[1ls8 | + | <StructureSection load='1ls8' size='340' side='right'caption='[[1ls8]]' scene=''> |
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1ls8]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1ls8]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Bombyx_mori Bombyx mori]. Full experimental information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1LS8 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1LS8 FirstGlance]. <br> |
| - | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ls8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ls8 OCA], [https://pdbe.org/1ls8 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ls8 RCSB], [https://www.ebi.ac.uk/pdbsum/1ls8 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ls8 ProSAT]</span></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">Solution NMR</td></tr> |
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ls8 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ls8 OCA], [https://pdbe.org/1ls8 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ls8 RCSB], [https://www.ebi.ac.uk/pdbsum/1ls8 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ls8 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/PBP_BOMMO PBP_BOMMO] This major soluble protein in olfactory sensilla of male moths serves to solubilize the extremely hydrophobic pheromone molecules such as bombykol and to transport pheromone through the aqueous lymph to receptors located on olfactory cilia. | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 18: | Line 19: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ls8 ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ls8 ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The nuclear magnetic resonance structure of the unliganded pheromone-binding protein (PBP) from Bombyx mori at pH above 6.5, BmPBP(B), consists of seven helices with residues 3-8, 16-22, 29-32, 46-59, 70-79, 84-100, and 107-124, and contains the three disulfide bridges 19-54, 50-108, and 97-117. This polypeptide fold encloses a large hydrophobic cavity, with a sufficient volume to accommodate the natural ligand bombykol. The polypeptide folds in free BmPBP(B) and in crystals of a BmPBP-bombykol complex are nearly identical, indicating that the B-form of BmPBP in solution represents the active conformation for ligand binding. | ||
| - | |||
| - | NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH.,Lee D, Damberger FF, Peng G, Horst R, Guntert P, Nikonova L, Leal WS, Wuthrich K FEBS Lett. 2002 Nov 6;531(2):314-8. PMID:12417333<ref>PMID:12417333</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1ls8" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Odorant binding protein|Odorant binding protein]] | *[[Odorant binding protein|Odorant binding protein]] | ||
| - | == References == | ||
| - | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Bombyx mori]] |
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Damberger | + | [[Category: Damberger F]] |
| - | [[Category: Guntert | + | [[Category: Guntert P]] |
| - | [[Category: Horst | + | [[Category: Horst R]] |
| - | [[Category: Leal | + | [[Category: Leal WS]] |
| - | [[Category: Lee | + | [[Category: Lee D]] |
| - | [[Category: Wuthrich | + | [[Category: Wuthrich K]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Revision as of 08:27, 10 April 2024
NMR structure of the unliganded Bombyx mori pheromone-binding protein at physiological pH
| |||||||||||
Categories: Bombyx mori | Large Structures | Damberger F | Guntert P | Horst R | Leal WS | Lee D | Wuthrich K