We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1diz
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1diz' size='340' side='right'caption='[[1diz]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='1diz' size='340' side='right'caption='[[1diz]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
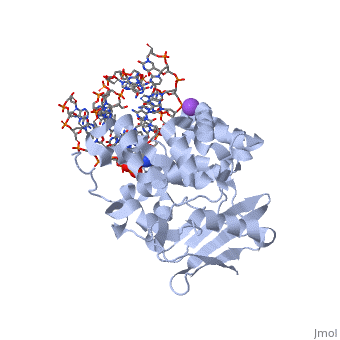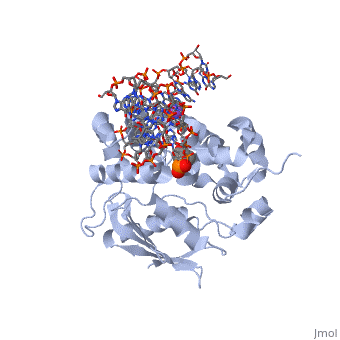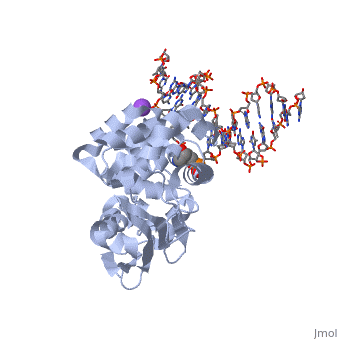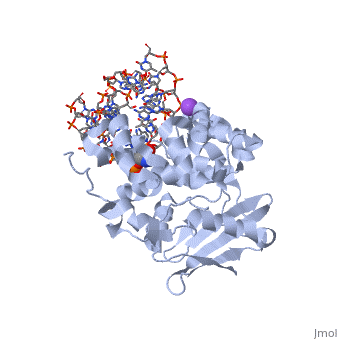
| - | <table><tr><td colspan='2'>[[1diz]] is a 6 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1diz]] is a 6 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1DIZ OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1DIZ FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=NA:SODIUM+ION'>NA</scene>, <scene name='pdbligand=NRI:PHOSPHORIC+ACID+MONO-(4-HYDROXY-PYRROLIDIN-3-YLMETHYL)+ESTER'>NRI</scene></td></tr> |
| - | + | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1diz FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1diz OCA], [https://pdbe.org/1diz PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1diz RCSB], [https://www.ebi.ac.uk/pdbsum/1diz PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1diz ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1diz FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1diz OCA], [https://pdbe.org/1diz PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1diz RCSB], [https://www.ebi.ac.uk/pdbsum/1diz PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1diz ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/3MG2_ECOLI 3MG2_ECOLI] Hydrolysis of the deoxyribose N-glycosidic bond to excise 3-methyladenine, 3-methylguanine, 7-methylguanine, O2-methylthymine, and O2-methylcytosine from the damaged DNA polymer formed by alkylation lesions. | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 21: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1diz ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1diz ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The Escherichia coli AlkA protein is a base excision repair glycosylase that removes a variety of alkylated bases from DNA. The 2.5 A crystal structure of AlkA complexed to DNA shows a large distortion in the bound DNA. The enzyme flips a 1-azaribose abasic nucleotide out of DNA and induces a 66 degrees bend in the DNA with a marked widening of the minor groove. The position of the 1-azaribose in the enzyme active site suggests an S(N)1-type mechanism for the glycosylase reaction, in which the essential catalytic Asp238 provides direct assistance for base removal. Catalytic selectivity might result from the enhanced stacking of positively charged, alkylated bases against the aromatic side chain of Trp272 in conjunction with the relative ease of cleaving the weakened glycosylic bond of these modified nucleotides. The structure of the AlkA-DNA complex offers the first glimpse of a helix-hairpin-helix (HhH) glycosylase complexed to DNA. Modeling studies suggest that other HhH glycosylases can bind to DNA in a similar manner. | ||
| - | |||
| - | DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA.,Hollis T, Ichikawa Y, Ellenberger T EMBO J. 2000 Feb 15;19(4):758-66. PMID:10675345<ref>PMID:10675345</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1diz" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[DNA glycosylase 3D structures|DNA glycosylase 3D structures]] | *[[DNA glycosylase 3D structures|DNA glycosylase 3D structures]] | ||
| - | == References == | ||
| - | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Escherichia coli]] |
| - | + | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: Ellenberger | + | [[Category: Ellenberger TE]] |
| - | [[Category: Hollis | + | [[Category: Hollis T]] |
| - | [[Category: Ichikawa | + | [[Category: Ichikawa Y]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
CRYSTAL STRUCTURE OF E. COLI 3-METHYLADENINE DNA GLYCOSYLASE (ALKA) COMPLEXED WITH DNA
| |||||||||||