We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Transmembrane protease serine 2
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
== Protease activity == | == Protease activity == | ||
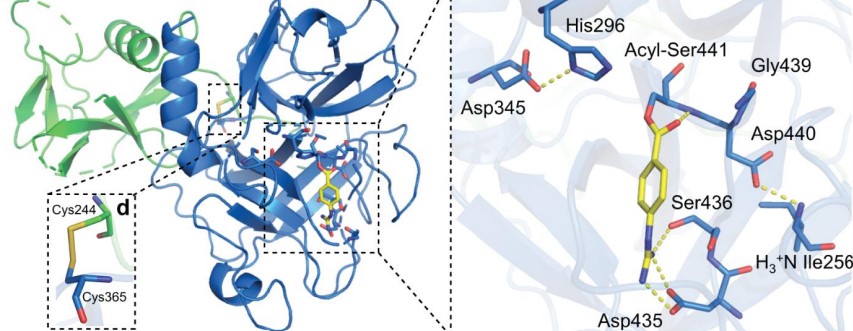
| - | + | TMPRSS2, as a serine protease, cleaves peptide bonds present after positively charged residues (lysine or arginine). The main player in the catalytic mechanism is the catalytic triad formed by His296, Asp345, and Ser441. This three aminoacids are located in the active site of the enzyme. <ref>DOI 10.1073/pnas.87.17.6659</ref> | |
The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | The substrate specificity is achieved with the presence of a negatively charged Asp residue at the bottom of a cavity usually indicated as “S1 specificity pocket”. <ref>DOI 10.1016/j.ejps.2020.105495</ref> | ||
| Line 44: | Line 44: | ||
=== Protein === | === Protein === | ||
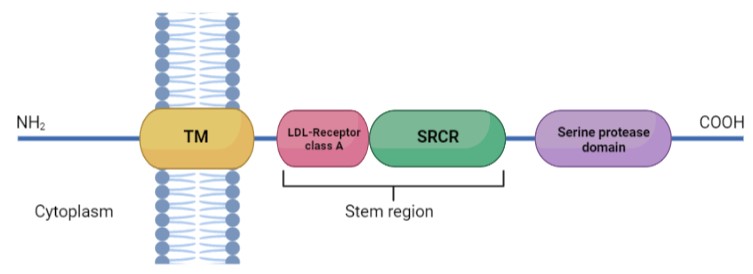
| - | TMPRSS2 is a 492 amino acid single-pass type II membrane protein. This protein is defined by the presence of an N-terminal cytoplasmic domain, a transmembrane helical domain, and three extracellular domains: <ref>DOI 10.1136/jclinpath-2020-206987</ref><scene name='89/897681/Domains/2'>TMPRSS2 protein domains</scene> | + | TMPRSS2 is a 492 amino acid single-pass type II membrane protein. This protein is defined by the presence of an N-terminal cytoplasmic domain, a transmembrane helical domain (aa 84-106), and three extracellular domains: <ref>DOI 10.1136/jclinpath-2020-206987</ref><scene name='89/897681/Domains/2'>TMPRSS2 protein domains</scene> |
| - | *Low-density lipoprotein (LDL)-receptor class A domain: which forms a binding site for calcium | + | *Low-density lipoprotein (LDL)-receptor class A domain: which forms a binding site for calcium. Expands from amino acid 113 to 148. |
| - | *Scavenger receptor cysteine-rich domain (SRCR) | + | *Scavenger receptor cysteine-rich domain (SRCR) of group A from amino acid 149 to 242. |
| - | *Peptidase S1 domain, also known as serine protease domain (SPD) | + | *Peptidase S1 domain, also known as serine protease domain (SPD) from amino acid 255 to 492. |
[[Image:TMPRSS2.jpg]] | [[Image:TMPRSS2.jpg]] | ||
Revision as of 15:03, 30 November 2021
TMPRSS2 is a membrane protein belonging to the type II transmembrane serine protease (TTSP) family. It is functionally classified as a trypsin-like protease (TLP). [1] Serine proteases are known to be involved in many physiological and pathological processes.
| |||||||||||
References
- ↑ Sgrignani J, Cavalli A. Computational Identification of a Putative Allosteric Binding Pocket in TMPRSS2. Front Mol Biosci. 2021 Apr 30;8:666626. doi: 10.3389/fmolb.2021.666626., eCollection 2021. PMID:33996911 doi:http://dx.doi.org/10.3389/fmolb.2021.666626
- ↑ Evnin LB, Vasquez JR, Craik CS. Substrate specificity of trypsin investigated by using a genetic selection. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6659-63. doi: 10.1073/pnas.87.17.6659. PMID:2204062 doi:http://dx.doi.org/10.1073/pnas.87.17.6659
- ↑ Singh N, Decroly E, Khatib AM, Villoutreix BO. Structure-based drug repositioning over the human TMPRSS2 protease domain: search for chemical probes able to repress SARS-CoV-2 Spike protein cleavages. Eur J Pharm Sci. 2020 Oct 1;153:105495. doi: 10.1016/j.ejps.2020.105495. Epub, 2020 Jul 28. PMID:32730844 doi:http://dx.doi.org/10.1016/j.ejps.2020.105495
- ↑ Lam DK, Dang D, Flynn AN, Hardt M, Schmidt BL. TMPRSS2, a novel membrane-anchored mediator in cancer pain. Pain. 2015 May;156(5):923-930. doi: 10.1097/j.pain.0000000000000130. PMID:25734995 doi:http://dx.doi.org/10.1097/j.pain.0000000000000130
- ↑ St John J, Powell K, Conley-Lacomb MK, Chinni SR. TMPRSS2-ERG Fusion Gene Expression in Prostate Tumor Cells and Its Clinical and Biological Significance in Prostate Cancer Progression. J Cancer Sci Ther. 2012 Apr 26;4(4):94-101. doi: 10.4172/1948-5956.1000119. PMID:23264855 doi:http://dx.doi.org/10.4172/1948-5956.1000119
- ↑ Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005 Oct 28;310(5748):644-8. doi: 10.1126/science.1117679. PMID:16254181 doi:http://dx.doi.org/10.1126/science.1117679
- ↑ Carrere S, Verger A, Flourens A, Stehelin D, Duterque-Coquillaud M. Erg proteins, transcription factors of the Ets family, form homo, heterodimers and ternary complexes via two distinct domains. Oncogene. 1998 Jun 25;16(25):3261-8. doi: 10.1038/sj.onc.1201868. PMID:9681824 doi:http://dx.doi.org/10.1038/sj.onc.1201868
- ↑ Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010 May 18;17(5):443-54. doi: 10.1016/j.ccr.2010.03.018. PMID:20478527 doi:http://dx.doi.org/10.1016/j.ccr.2010.03.018
- ↑ Farooqi AA, Hou MF, Chen CC, Wang CL, Chang HW. Androgen receptor and gene network: Micromechanics reassemble the signaling machinery of TMPRSS2-ERG positive prostate cancer cells. Cancer Cell Int. 2014 Apr 17;14:34. doi: 10.1186/1475-2867-14-34. eCollection, 2014. PMID:24739220 doi:http://dx.doi.org/10.1186/1475-2867-14-34
- ↑ Thunders M, Delahunt B. Gene of the month: TMPRSS2 (transmembrane serine protease 2). J Clin Pathol. 2020 Dec;73(12):773-776. doi: 10.1136/jclinpath-2020-206987. Epub , 2020 Sep 1. PMID:32873700 doi:http://dx.doi.org/10.1136/jclinpath-2020-206987
- ↑ Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, Raitano AB, Jakobovits A. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001 Feb 15;61(4):1686-92. PMID:11245484
- ↑ Bertram S, Glowacka I, Blazejewska P, Soilleux E, Allen P, Danisch S, Steffen I, Choi SY, Park Y, Schneider H, Schughart K, Pohlmann S. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol. 2010 Oct;84(19):10016-25. doi: 10.1128/JVI.00239-10. Epub 2010 Jul 14. PMID:20631123 doi:http://dx.doi.org/10.1128/JVI.00239-10
- ↑ Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: A potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017 Nov;142:1-10. doi: 10.1016/j.biochi.2017.07.016. Epub 2017 Aug 1. PMID:28778717 doi:http://dx.doi.org/10.1016/j.biochi.2017.07.016
- ↑ Amraei R, Rahimi N. COVID-19, Renin-Angiotensin System and Endothelial Dysfunction. Cells. 2020 Jul 9;9(7). pii: cells9071652. doi: 10.3390/cells9071652. PMID:32660065 doi:http://dx.doi.org/10.3390/cells9071652
- ↑ doi: https://dx.doi.org/10.1101/2021.06.23.449282
- ↑ Shrimp JH, Kales SC, Sanderson PE, Simeonov A, Shen M, Hall MD. An Enzymatic TMPRSS2 Assay for Assessment of Clinical Candidates and Discovery of Inhibitors as Potential Treatment of COVID-19. bioRxiv. 2020 Aug 6. doi: 10.1101/2020.06.23.167544. PMID:32596694 doi:http://dx.doi.org/10.1101/2020.06.23.167544
- ↑ Hitomi Y, Ikari N, Fujii S. Inhibitory effect of a new synthetic protease inhibitor (FUT-175) on the coagulation system. Haemostasis. 1985;15(3):164-8. doi: 10.1159/000215139. PMID:3161808 doi:http://dx.doi.org/10.1159/000215139
- ↑ Maggio R, Corsini GU. Repurposing the mucolytic cough suppressant and TMPRSS2 protease inhibitor bromhexine for the prevention and management of SARS-CoV-2 infection. Pharmacol Res. 2020 Jul;157:104837. doi: 10.1016/j.phrs.2020.104837. Epub 2020, Apr 22. PMID:32334052 doi:http://dx.doi.org/10.1016/j.phrs.2020.104837
- ↑ doi: https://dx.doi.org/10.20944/preprints202003.0360.v2
Proteopedia Page Contributors and Editors (what is this?)
Paula R. Mallavibarrena, Ines Muniesa-Martinez, Michal Harel, Laura Aleixos Juan, Jaime Prilusky