We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1697
From Proteopedia
(Difference between revisions)
| Line 12: | Line 12: | ||
== Important amino acids== | == Important amino acids== | ||
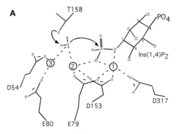
| - | In the protein the | + | In the protein the substrate is <scene name='89/892740/Ligand_view/1'>D-MYO-INOSITOL-1,4-BISPHOSPHATE</scene>, will be dephosphorylated by the water brought in by the magnesium ions.In this protein the amino acids do not interact with the substrate itself, instead they interact with the metal ions. When magnesium is present, and the protein therefore activated, the amino acids will hold onto the magnesium ions. These magnesium ions that interact with the inositol and water so that it can dephosphorylate the substrate. The motif of enzymes in this family is DPIDXT. In this particular protein the motif is <scene name='89/892740/Motif_view/1'>D54, E80, E79, D153, and D317</scene>. There are some mutations in the enzyme but the overall function of the protein is still the same. |
== Structural highlights == | == Structural highlights == | ||
| - | The protein has thirteen structural elements. The amino acids in the bind to the substrate all participate in hydrogen bonding to the substrate | + | The protein has thirteen structural elements. The amino acids in the bind to the substrate all participate in hydrogen bonding to the substrate. The structure is <scene name='89/892740/Secondary_structures/1'>77% helix and 23% beta sheet</scene>, the betta sheets allow for twisting of the molecule so that the substrate better fits within the enzyme. The helices allow for hydrogen bonding throughout to stabilize the structure with assistance from metal ions; specifically magnesium ions. <ref> PMID 33172890 </ref> |
There is no quaternary structures in the protein but several <scene name='89/892740/7kir_quaternary_structures/1'>tertiary structures</scene> that play an important role in the structure of the protein and therefore its function. There are many covalent bonds due to hydrophobic and nonpolar amino acids that form the structure. The amino acids involved with the <scene name='89/892740/Quat_structure_catalytic_amino/1'>catalytic amino acids are mostly polar</scene>, the magnesium ions assist in pulling the enzyme into shape that allow for it to be in the <scene name='89/892740/Spacefill_7kir/1'>proper shape</scene> so that it can dephosphorylate the inositol. It wraps around the magnesium ions so that it can better attach to the water and then attack the phosphate on inositol. When the lithium ions interact with the enzyme a shape change occurs that will not allow for the magnesium ions to attach to the enzyme, and therefore the water molecules. | There is no quaternary structures in the protein but several <scene name='89/892740/7kir_quaternary_structures/1'>tertiary structures</scene> that play an important role in the structure of the protein and therefore its function. There are many covalent bonds due to hydrophobic and nonpolar amino acids that form the structure. The amino acids involved with the <scene name='89/892740/Quat_structure_catalytic_amino/1'>catalytic amino acids are mostly polar</scene>, the magnesium ions assist in pulling the enzyme into shape that allow for it to be in the <scene name='89/892740/Spacefill_7kir/1'>proper shape</scene> so that it can dephosphorylate the inositol. It wraps around the magnesium ions so that it can better attach to the water and then attack the phosphate on inositol. When the lithium ions interact with the enzyme a shape change occurs that will not allow for the magnesium ions to attach to the enzyme, and therefore the water molecules. | ||
The rest of the enzyme is circled around the substrate which allows for a better ability to bind to the substrate, it is shown here how the <scene name='89/892740/Hydrophobic_effect/1'>hydrophobic effect</scene> how the nonpolar and polar amino acids allow for it to have this shape. | The rest of the enzyme is circled around the substrate which allows for a better ability to bind to the substrate, it is shown here how the <scene name='89/892740/Hydrophobic_effect/1'>hydrophobic effect</scene> how the nonpolar and polar amino acids allow for it to have this shape. | ||
Revision as of 03:32, 9 December 2021
| This Sandbox is Reserved from 10/01/2021 through 01/01//2022 for use in Biochemistry taught by Bonnie Hall at Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1690 through Sandbox Reserved 1699. |
To get started:
More help: Help:Editing |
7KIR
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Dollins DE, Xiong JP, Endo-Streeter S, Anderson DE, Bansal VS, Ponder JW, Ren Y, York JD. A Structural Basis for Lithium and Substrate Binding of an Inositide Phosphatase. J Biol Chem. 2020 Nov 10. pii: RA120.014057. doi: 10.1074/jbc.RA120.014057. PMID:33172890 doi:http://dx.doi.org/10.1074/jbc.RA120.014057