User:Natalia de Araujo./Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
---- | ---- | ||
| - | '''The first structure''', displayed on the left-hand side of the screen, corresponds to [[4jon]], the Crystal structure of CEP170's transcript variant beta, from Homo sapiens. 4jon is a 5 chain structure obtained by ''X-ray Crystallography'' [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions.&text=In%20a%20single%2Dcrystal%20X,is%20mounted%20on%20a%20goniometer.] and represents the first 126 residues of CEP170's N-termini region. | + | '''The first structure''', displayed on the left-hand side of the screen, corresponds to [[4jon]], the Crystal structure of CEP170's transcript variant beta, from Homo sapiens. 4jon is a <scene name='89/897701/4jon_5_chain_structure/1'>5 chain structure</scene> obtained by ''X-ray Crystallography'' [https://en.wikipedia.org/wiki/X-ray_crystallography#:~:text=X%2Dray%20crystallography%20is%20the,diffract%20into%20many%20specific%20directions.&text=In%20a%20single%2Dcrystal%20X,is%20mounted%20on%20a%20goniometer.] and represents the first 126 residues of CEP170's N-termini region. |
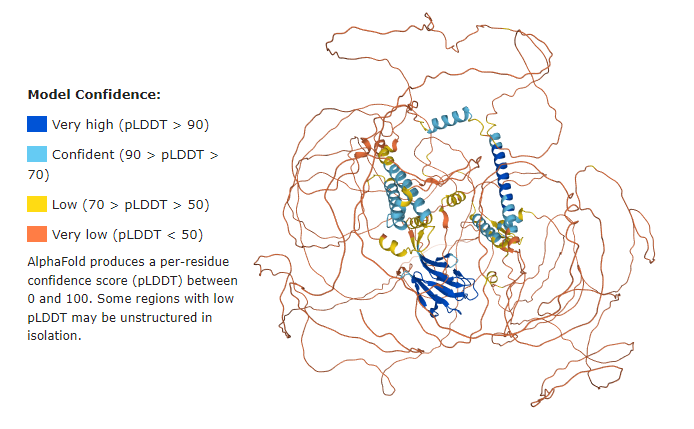
'''The second structure''' available for CEP170 is <scene name='89/897701/Q5sw79/1'>Q5SW79</scene>, a ''predicted'' 3D model representing its full lenght (1-1584). | '''The second structure''' available for CEP170 is <scene name='89/897701/Q5sw79/1'>Q5SW79</scene>, a ''predicted'' 3D model representing its full lenght (1-1584). | ||
Revision as of 10:21, 13 December 2021
Centrosomal protein of 170kDa (CEP170)
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005 Mar;16(3):1095-107. Epub 2004 Dec 22. PMID:15616186 doi:E04-10-0939
- ↑ Gu C, Wang W, Tang X, Xu T, Zhang Y, Guo M, Wei R, Wang Y, Jurczyszyn A, Janz S, Beksac M, Zhan F, Seckinger A, Hose D, Pan J, Yang Y. CHEK1 and circCHEK1_246aa evoke chromosomal instability and induce bone lesion formation in multiple myeloma. Mol Cancer. 2021 Jun 5;20(1):84. doi: 10.1186/s12943-021-01380-0. PMID:34090465 doi:http://dx.doi.org/10.1186/s12943-021-01380-0
- ↑ Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005 Mar;16(3):1095-107. Epub 2004 Dec 22. PMID:15616186 doi:E04-10-0939
- ↑ Welburn JP, Cheeseman IM. The microtubule-binding protein Cep170 promotes the targeting of the kinesin-13 depolymerase Kif2b to the mitotic spindle. Mol Biol Cell. 2012 Dec;23(24):4786-95. doi: 10.1091/mbc.E12-03-0214. Epub 2012, Oct 19. PMID:23087211 doi:http://dx.doi.org/10.1091/mbc.E12-03-0214
- ↑ Pillai S, Nguyen J, Johnson J, Haura E, Coppola D, Chellappan S. Tank binding kinase 1 is a centrosome-associated kinase necessary for microtubule dynamics and mitosis. Nat Commun. 2015 Dec 10;6:10072. doi: 10.1038/ncomms10072. PMID:26656453 doi:http://dx.doi.org/10.1038/ncomms10072
- ↑ Huang N, Xia Y, Zhang D, Wang S, Bao Y, He R, Teng J, Chen J. Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68. Nat Commun. 2017 Apr 19;8:15057. doi: 10.1038/ncomms15057. PMID:28422092 doi:http://dx.doi.org/10.1038/ncomms15057
- ↑ Barenz F, Kschonsak YT, Meyer A, Jafarpour A, Lorenz H, Hoffmann I. Ccdc61 controls centrosomal localization of Cep170 and is required for spindle assembly and symmetry. Mol Biol Cell. 2018 Dec 15;29(26):3105-3118. doi: 10.1091/mbc.E18-02-0115. Epub, 2018 Oct 24. PMID:30354798 doi:http://dx.doi.org/10.1091/mbc.E18-02-0115
- ↑ Qin Y, Zhou Y, Shen Z, Xu B, Chen M, Li Y, Chen M, Behrens A, Zhou J, Qi X, Meng W, Wang Y, Gao F. WDR62 is involved in spindle assembly by interacting with CEP170 in spermatogenesis. Development. 2019 Oct 21;146(20). pii: dev.174128. doi: 10.1242/dev.174128. PMID:31533924 doi:http://dx.doi.org/10.1242/dev.174128
- ↑ Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005 Mar;16(3):1095-107. Epub 2004 Dec 22. PMID:15616186 doi:E04-10-0939
- ↑ Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005 Mar;16(3):1095-107. Epub 2004 Dec 22. PMID:15616186 doi:E04-10-0939
- ↑ Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005 Mar;16(3):1095-107. Epub 2004 Dec 22. PMID:15616186 doi:E04-10-0939