Sandbox reserved 1651
From Proteopedia
(Difference between revisions)
| Line 28: | Line 28: | ||
=== Regulation of the product length === | === Regulation of the product length === | ||
| - | The hydrophobic tunnel of DHDDS is formed by 2 α-helix and 4 β-strands. The opening between the 2 α-helices is larger in DHDDS compared to short and medium-chain cis-PT. | + | The hydrophobic tunnel of DHDDS is formed by 2 α-helix and 4 β-strands. The opening between the 2 α-helices is larger in DHDDS compared to short and medium-chain cis-PT. Larger is the diameter, better is the accommodation of a longer product. |
| Line 38: | Line 38: | ||
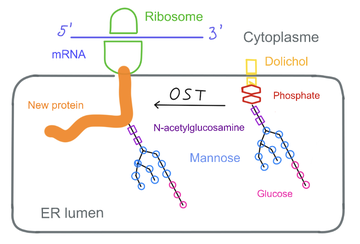
The [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]is a post-translational modification realized in the endoplasmic reticulum of the cell. This process consists in linking a [https://en.wikipedia.org/wiki/Glycan glycan] to a protein, which provide the biological protein fonction and [https://en.wikipedia.org/wiki/Protein_folding folding]. The hcis-PT produces DHDD (dehydrodolichyl diphosphate), an important precursor molecule for the dolichol-phosphate lipid carrier needed in the [https://fr.wikipedia.org/wiki/N-glycosylation N-glycosylation] reaction. | The [https://en.wikipedia.org/wiki/N-linked_glycosylation N-glycosylation]is a post-translational modification realized in the endoplasmic reticulum of the cell. This process consists in linking a [https://en.wikipedia.org/wiki/Glycan glycan] to a protein, which provide the biological protein fonction and [https://en.wikipedia.org/wiki/Protein_folding folding]. The hcis-PT produces DHDD (dehydrodolichyl diphosphate), an important precursor molecule for the dolichol-phosphate lipid carrier needed in the [https://fr.wikipedia.org/wiki/N-glycosylation N-glycosylation] reaction. | ||
| - | [[image:N-glycosylation.png | thumb | center|360px| upright=10|'''N-glycosylation''']] | + | [[image:N-glycosylation.png | thumb | center|360px| upright=10|'''N-glycosylation''']] [2] |
=== Disease comprehension === | === Disease comprehension === | ||
| - | The [https://en.wikipedia.org/wiki/Retinitis_pigmentosa Retinitis pigmentosa] is a [https://en.wikipedia.org/wiki/Genetic_disorder hereditary disease]. | + | The [https://en.wikipedia.org/wiki/Retinitis_pigmentosa Retinitis pigmentosa] is a [https://en.wikipedia.org/wiki/Genetic_disorder hereditary disease]. Patients lose progressively parts of their sight: night vision as a teenager, side vision as a young adult, and central vision in later life. This is due to the progressive decrease of [https://en.wikipedia.org/wiki/Rod_cell rod] and [https://en.wikipedia.org/wiki/Cone_cell cone] [https://en.wikipedia.org/wiki/Photoreceptor_cell photoreceptor cells] which provide colored vision. Scientists observed that symptoms appear many years after the beginning of the photoreceptor’s [https://en.wikipedia.org/wiki/Degenerative_disease degeneration] in most cases. Retinitis pigmentosa can be encoded by many genes. More than 45 have been identified but these genes concern only 60% of sick patients. Therefore 40% of cases of Retinitis pigmentosa are unidentified genes. Until today there is no cure. However, studies showed a slow down of the disease with the intake of [https://en.wikipedia.org/wiki/Vitamin_A vitamin A], [https://en.wikipedia.org/wiki/Palmitic_acid palmitate] foods and [https://en.wikipedia.org/wiki/Omega-3_fatty_acid omega-3]-rich fish [3]. |
For instance, <scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene> missense mutations can provoke an [https://en.wikipedia.org/w/index.php?title=Autosomal_Recessive&redirect=no autosomal recessive] Retinitis pigmentosa (arRP). These mutations concern the S1 and S2 sites of the active site where pyrophosphate can bind and most of the mutations related to the disease impact directly the substrate binding, according to scientists. For one mutation, <scene name='87/872232/K42/3'>K42E</scene>, it is more complicated. Scientists remarked that, in the <scene name='87/872232/Salt_bridge_k42-e234/1'>wild type, R38 points toward the active-site cavity, while K42 and E234 form a stable salt bridge</scene> (short distance). The mutation K42E provokes in the protein scale, hypothetically, an interaction with the adjacent active-site residues R38 positively charged. So R38 points away from the active site cavity, forming a new stable [[salt bridge]] with the mutant E42. The distance between K42E and E234 is longer due to the charge repulsion. Finally, experiments proved that aberrant polar networks are due to the K42E mutation, disturbing the active-site residues which can’t interact with the substrate and leading to a decrease of the catalytic activity [1]. | For instance, <scene name='87/872232/Subunit_dhdds/1'>DHDDS</scene> missense mutations can provoke an [https://en.wikipedia.org/w/index.php?title=Autosomal_Recessive&redirect=no autosomal recessive] Retinitis pigmentosa (arRP). These mutations concern the S1 and S2 sites of the active site where pyrophosphate can bind and most of the mutations related to the disease impact directly the substrate binding, according to scientists. For one mutation, <scene name='87/872232/K42/3'>K42E</scene>, it is more complicated. Scientists remarked that, in the <scene name='87/872232/Salt_bridge_k42-e234/1'>wild type, R38 points toward the active-site cavity, while K42 and E234 form a stable salt bridge</scene> (short distance). The mutation K42E provokes in the protein scale, hypothetically, an interaction with the adjacent active-site residues R38 positively charged. So R38 points away from the active site cavity, forming a new stable [[salt bridge]] with the mutant E42. The distance between K42E and E234 is longer due to the charge repulsion. Finally, experiments proved that aberrant polar networks are due to the K42E mutation, disturbing the active-site residues which can’t interact with the substrate and leading to a decrease of the catalytic activity [1]. | ||
| Line 53: | Line 53: | ||
[1] Michal Lisnyansky Bar-El et al. «Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase ». ''Nature Communications''. '''11''':523, (2020). | [1] Michal Lisnyansky Bar-El et al. «Structural basis of heterotetrameric assembly and disease mutations in the human cis-prenyltransferase ». ''Nature Communications''. '''11''':523, (2020). | ||
| - | [2 ]Dyonne T Hartong et al. « Retinitis Pigmentosa ». ''The Lancet''. 18;368(9549):1795‑809, (2006) | + | [2]BAR-EL, Michal Lisnyansky, VANKOVA, Pavla, MAN, Petr, et al. Structure of the human heterotetrameric cis-prenyltransferase complex. bioRxiv, 2020. |
| + | |||
| + | [3 ]Dyonne T Hartong et al. « Retinitis Pigmentosa ». ''The Lancet''. 18;368(9549):1795‑809, (2006) | ||
Revision as of 17:58, 19 January 2022
Heterotetrameric Cis-Prenyltransferase Complex
| |||||||||||