We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1660
From Proteopedia
(Difference between revisions)
| Line 13: | Line 13: | ||
__TOC__ | __TOC__ | ||
| - | == Function == | + | =='''Function'''== |
[[Image:CD1d dependent lipid antigen presentation.jpg | thumb ||left|190px|'''CD1d dependent lipid antigen presentation.''']] | [[Image:CD1d dependent lipid antigen presentation.jpg | thumb ||left|190px|'''CD1d dependent lipid antigen presentation.''']] | ||
| Line 22: | Line 22: | ||
'''CD1d protein''' is a molecule of the [https://en.wikipedia.org/wiki/Immune_system immune system], involved in the presentation of a lipid antigen to NKT cells, which are a subset of [https://en.wikipedia.org/wiki/T_cell T cells]. Indeed, these proteins are located on the plasma membrane of [https://en.wikipedia.org/wiki/Antigen-presenting_cell APC cells]. When the recognition between the CD1d bound to its lipid ligand and the [[TCR]] of a NKT cell occurs, the [https://en.wikipedia.org/wiki/Lymphocyte lymphocyte] turns out to be activated. Thus, the production of cytotoxic molecules such as [[interleukin]]-4 and [https://en.wikipedia.org/wiki/Interferon_gamma IFN-gamma] is triggered by this activation, leading to a Th2 or Th1 immune response respectively<ref name="properties">Joyce, S. CD1d and Natural T Cells: How Their Properties Jump-Start the Immune System. CMLS, Cell. Mol. Life Sci. 2001, 58 (3), 442–469. https://doi.org/10.1007/PL00000869.</ref><ref name="affinity">Rossjohn, J., Pellicci, D. G., Patel, O., Gapin, L., & Godfrey, D. I. (2012). Recognition of CD1d-restricted antigens by natural killer T cells. Nature reviews. Immunology, 12(12), 845–857. https://doi.org/10.1038/nri3328.</ref>. Therefore, CD1d proteins are precursors of the adaptive immune reaction. As a result, a deficiency of CD1d protein may lead to a deficiency of the NKT cells functioning and thus to [https://en.wikipedia.org/wiki/Autoimmune_disease autoimmune diseases] and [https://en.wikipedia.org/wiki/Cancer cancers]. | '''CD1d protein''' is a molecule of the [https://en.wikipedia.org/wiki/Immune_system immune system], involved in the presentation of a lipid antigen to NKT cells, which are a subset of [https://en.wikipedia.org/wiki/T_cell T cells]. Indeed, these proteins are located on the plasma membrane of [https://en.wikipedia.org/wiki/Antigen-presenting_cell APC cells]. When the recognition between the CD1d bound to its lipid ligand and the [[TCR]] of a NKT cell occurs, the [https://en.wikipedia.org/wiki/Lymphocyte lymphocyte] turns out to be activated. Thus, the production of cytotoxic molecules such as [[interleukin]]-4 and [https://en.wikipedia.org/wiki/Interferon_gamma IFN-gamma] is triggered by this activation, leading to a Th2 or Th1 immune response respectively<ref name="properties">Joyce, S. CD1d and Natural T Cells: How Their Properties Jump-Start the Immune System. CMLS, Cell. Mol. Life Sci. 2001, 58 (3), 442–469. https://doi.org/10.1007/PL00000869.</ref><ref name="affinity">Rossjohn, J., Pellicci, D. G., Patel, O., Gapin, L., & Godfrey, D. I. (2012). Recognition of CD1d-restricted antigens by natural killer T cells. Nature reviews. Immunology, 12(12), 845–857. https://doi.org/10.1038/nri3328.</ref>. Therefore, CD1d proteins are precursors of the adaptive immune reaction. As a result, a deficiency of CD1d protein may lead to a deficiency of the NKT cells functioning and thus to [https://en.wikipedia.org/wiki/Autoimmune_disease autoimmune diseases] and [https://en.wikipedia.org/wiki/Cancer cancers]. | ||
</p> | </p> | ||
| - | <br> | ||
==== Ligands presented by CD1d ==== | ==== Ligands presented by CD1d ==== | ||
| Line 31: | Line 30: | ||
<br> | <br> | ||
| - | == Structure of mouse CD1d == | + | =='''Structure of mouse CD1d'''== |
<p align="justify"> | <p align="justify"> | ||
| Line 38: | Line 37: | ||
:- a <scene name='86/868193/Beta-2-microglobulin_chain/7'>beta-2-microglobulin chain</scene> of 99 amino acids. <br> | :- a <scene name='86/868193/Beta-2-microglobulin_chain/7'>beta-2-microglobulin chain</scene> of 99 amino acids. <br> | ||
The alpha chain is made of three domains: alpha 1, alpha 2 and alpha 3. The association of alpha 1 and alpha 2 is composed of two [https://en.wikipedia.org/wiki/Beta_sheet beta-sheets] and a set of 2 [https://proteopedia.org/wiki/index.php/Alpha_helix alpha helixes]. Each beta-sheet contains four antiparallel strands. The ligand binds between the two alpha 1 and 2 helices. The alpha 3 domain is non-covalently bound with beta-2-microglobulin domain. | The alpha chain is made of three domains: alpha 1, alpha 2 and alpha 3. The association of alpha 1 and alpha 2 is composed of two [https://en.wikipedia.org/wiki/Beta_sheet beta-sheets] and a set of 2 [https://proteopedia.org/wiki/index.php/Alpha_helix alpha helixes]. Each beta-sheet contains four antiparallel strands. The ligand binds between the two alpha 1 and 2 helices. The alpha 3 domain is non-covalently bound with beta-2-microglobulin domain. | ||
| - | Additionally, there are five <scene name='86/868193/Oligosaccharides/3'>oligosaccharides</scene><ref name="Structure"/> bound to the alpha chain via N-glycosylations, three of which have been clearly identified<ref name="oligo">Sriram, V., Willard, C.A., Liu, J., & Brutkiewicz, R.R.(2008). Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology, 123:272–281.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2433293/</ref>. The total molecular weight of the alpha chain is 33 kDa when not associated to any oligosaccharide and 55 kDa when oligosaccharides are associated to the chain<ref name="properties"/>.< | + | Additionally, there are five <scene name='86/868193/Oligosaccharides/3'>oligosaccharides</scene><ref name="Structure"/> bound to the alpha chain via N-glycosylations, three of which have been clearly identified<ref name="oligo">Sriram, V., Willard, C.A., Liu, J., & Brutkiewicz, R.R.(2008). Importance of N-linked glycosylation in the functional expression of murine CD1d1. Immunology, 123:272–281.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2433293/</ref>. The total molecular weight of the alpha chain is 33 kDa when not associated to any oligosaccharide and 55 kDa when oligosaccharides are associated to the chain<ref name="properties"/>.</p> |
CD1d molecules are structurally similar to [[Major Histocompatibility Complex Class I]], but present lipid antigens as opposed to peptides. Thus the cleft where the ligand can bind is different between MHC molecules and CD1d molecules. Indeed, the hydrophobic cleft of CD1d has a narrow opening.The recognition between the protein and its ligand occurs at a specific hydrophobic spot which creates an appropriate environment for the interaction to happen. This <scene name='86/868193/Site/3'>site</scene> is located at the A' and F' pockets in the region of the alpha helices<ref name="site">Schiefner, A.; Fujio, M.; Wu, D.; Wong, C.-H.; Wilson, I. A. Structural Evaluation of Potent NKT-Cell Agonists: Implications for Design of Novel Stimulatory Ligands. J Mol Biol 2009, 394 (1), 71–82. https://doi.org/10.1016/j.jmb.2009.08.061</ref>. | CD1d molecules are structurally similar to [[Major Histocompatibility Complex Class I]], but present lipid antigens as opposed to peptides. Thus the cleft where the ligand can bind is different between MHC molecules and CD1d molecules. Indeed, the hydrophobic cleft of CD1d has a narrow opening.The recognition between the protein and its ligand occurs at a specific hydrophobic spot which creates an appropriate environment for the interaction to happen. This <scene name='86/868193/Site/3'>site</scene> is located at the A' and F' pockets in the region of the alpha helices<ref name="site">Schiefner, A.; Fujio, M.; Wu, D.; Wong, C.-H.; Wilson, I. A. Structural Evaluation of Potent NKT-Cell Agonists: Implications for Design of Novel Stimulatory Ligands. J Mol Biol 2009, 394 (1), 71–82. https://doi.org/10.1016/j.jmb.2009.08.061</ref>. | ||
| - | < | + | <br><br> |
| - | + | =='''Impact of ligand-binding'''== | |
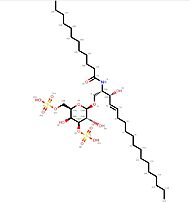
| - | == Impact of ligand-binding == | + | [[Image:C12DS.jpg | thumb ||left|190px|'''Ligand C12-di-sulfatide''']] |
| - | + | ||
==== Conformational variation ==== | ==== Conformational variation ==== | ||
| - | + | The stability of the CD1d-glycolipid complexes has an impact on the [https://en.wikipedia.org/wiki/Cytokine cytokine] release (cell signaling). Conformational variations that would stabilize the F’-pocket (primary site of interaction with the T cell receptor, NKT TCR) might increase CD1d affinity for the NKT TCR<ref name="affinity"/><ref name="site"/>. | |
| - | + | ||
| - | The stability of the CD1d-glycolipid complexes has an impact on the [https://en.wikipedia.org/wiki/Cytokine cytokine] release (cell signaling). Conformational variations that would stabilize the F’-pocket (primary site of interaction with the T cell receptor, NKT TCR) might increase CD1d affinity for the NKT TCR<ref name="affinity"/><ref name="site"/>. | + | |
| - | + | ||
The binding of a ligand (such as C12-di-sulfatide) leads to <scene name='86/868193/Conformational_variations/1'>conformational variations</scene> of CD1d molecule (The moving molecules are the oligosaccharides molecules ; the ligand is not represented). | The binding of a ligand (such as C12-di-sulfatide) leads to <scene name='86/868193/Conformational_variations/1'>conformational variations</scene> of CD1d molecule (The moving molecules are the oligosaccharides molecules ; the ligand is not represented). | ||
| Line 56: | Line 51: | ||
<p align="justify"> | <p align="justify"> | ||
CD1d proteins lipid recognition is based on the interaction of the protein with its ligand. Nevertheless, the interaction relies on two recognitions. The first one is the recognition of the head of the lipid and the second one is the recognition of the length of the molecule. Because there are more than one condition to fill in order to interact with CD1d proteins, the affinity for a lipid depends itself on a plurality of parameters which modulates it<ref name="affinity"/><ref name="site"/><ref>McCarthy, C.; Shepherd, D.; Fleire, S.; Stronge, V. S.; Koch, M.; Illarionov, P. A.; Bossi, G.; Salio, M.; Denkberg, G.; Reddington, F.; Tarlton, A.; Reddy, B. G.; Schmidt, R. R.; Reiter, Y.; Griffiths, G. M.; van der Merwe, P. A.; Besra, G. S.; Jones, E. Y.; Batista, F. D.; Cerundolo, V. The Length of Lipids Bound to Human CD1d Molecules Modulates the Affinity of NKT Cell TCR and the Threshold of NKT Cell Activation. J Exp Med 2007, 204 (5), 1131–1144. https://doi.org/10.1084/jem.20062342.</ref>. | CD1d proteins lipid recognition is based on the interaction of the protein with its ligand. Nevertheless, the interaction relies on two recognitions. The first one is the recognition of the head of the lipid and the second one is the recognition of the length of the molecule. Because there are more than one condition to fill in order to interact with CD1d proteins, the affinity for a lipid depends itself on a plurality of parameters which modulates it<ref name="affinity"/><ref name="site"/><ref>McCarthy, C.; Shepherd, D.; Fleire, S.; Stronge, V. S.; Koch, M.; Illarionov, P. A.; Bossi, G.; Salio, M.; Denkberg, G.; Reddington, F.; Tarlton, A.; Reddy, B. G.; Schmidt, R. R.; Reiter, Y.; Griffiths, G. M.; van der Merwe, P. A.; Besra, G. S.; Jones, E. Y.; Batista, F. D.; Cerundolo, V. The Length of Lipids Bound to Human CD1d Molecules Modulates the Affinity of NKT Cell TCR and the Threshold of NKT Cell Activation. J Exp Med 2007, 204 (5), 1131–1144. https://doi.org/10.1084/jem.20062342.</ref>. | ||
| - | </p> | + | </p><br> |
| - | == Applications == | + | =='''Applications'''== |
==== Immunotherapeutic tool ==== | ==== Immunotherapeutic tool ==== | ||
Revision as of 17:10, 19 January 2022
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
| |||||||||||