We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1711
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
| - | + | = Neurofibromin = | |
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | <StructureSection load='7pgr' size='340' side='right' caption='Closed conformation of Neurofibromin' scene='90/905640/Closedoverall/1'> | ||
| + | |||
| + | --- | ||
| + | |||
| + | [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7692384/] | ||
| + | |||
| + | [[Image:Open.jpg|400px|right|thumb|Figure_1]] | ||
| + | |||
| + | == Introduction == | ||
| + | <ref name="Bergoug">PMID:33121128</ref> | ||
| + | == Structure == | ||
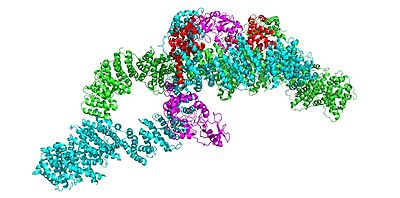
| + | Neurofibromin is a <scene name='90/905640/Homodimer/1'>homodimer</scene> with two identical chains (depicted as lime and cyan). | ||
| + | === Conformation: Open === | ||
| + | |||
| + | === Conformation: Closed === | ||
| + | In the overall closed <scene name='90/905640/Closedoverall/1'>closed neurofibromin</scene> and inactive conformation of the neurofibromin protein, both sets of the GRD and Sec14-PH domains are rotated in a way that they are inaccessible and inactive. The closed conformation has both protomers/chains in the closed structure, whereas the open conformation has one closed and one open protomer. You can see that in the closed conformation, Ras binding by the GRD domain is sterically hindered and there is no room for association with the Ras protein. | ||
| + | The closed conformation has the GRD and Sec14-PH domains oriented in a way that the amino acids C1032, H1558, and H1576 are in close proximity to each other to form a transition metal-binding site with zinc. The fourth coordination partner in this is water. | ||
| + | <scene name='90/905640/Closedoverall/6'>vertical view</scene> | ||
| + | |||
| + | === Domains === | ||
| + | The <scene name='90/905640/Grd_domains/2'>GRD domain</scene> of Neurofibromin, specifically the arginine finger (R1276), binds to the Ras + GTP complex. | ||
| + | === Key Players === | ||
| + | This is because an <scene name='90/905640/Arg_finger/1'>R1276</scene> (R1276) present in the GRD is critical for Ras binding and is only accessible when the GRD and Sec14-PH domains are rotated in such a way that there is no steric hindrance from the surrounding dimer chains. | ||
| + | Closed conformation is stabilized by one cysteine and two histidines that are coordinated with transition metal-binding sites with zinc. | ||
== Function == | == Function == | ||
| - | == | + | === Ras Control === |
| + | Ras is still promoting cell proliferation in this closed conformation because Neurofibromin is unable to hydrolyze Ras and inactivate it. In this open conformation, the Ras is not sterically hindered and the Arginine finger is accessible for Ras binding, thus allowing Neurofibromin to down-regulate Ras. | ||
| + | === Mutated === | ||
| - | == | + | == Diseases == |
| + | <ref name="Ransey">PMID:28504306</ref> | ||
| - | == Structural highlights == | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
| + | |||
| + | == Student Contributors == | ||
| + | *Hannah Luchinski | ||
Revision as of 19:10, 28 March 2022
Neurofibromin
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Hannah Luchinski