This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1717
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
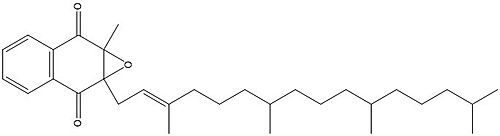
[[Image:Vitamin K epoxide.jpg|500 px|right|thumb|Figure 1. Vitamin K Epoxide structure]] | [[Image:Vitamin K epoxide.jpg|500 px|right|thumb|Figure 1. Vitamin K Epoxide structure]] | ||
| + | {{Template:CH462_Biochemistry_II_2022}}<!-- PLEASE ADD YOUR CONTENT BELOW HERE --> | ||
| + | ==Your Heading Here (maybe something like 'Structure')== | ||
| + | <StructureSection load='6wv3' size='340' side='right' caption='Caption for this structure' scene=''> | ||
| + | This is a default text for your page ''''''. Click above on '''edit this page''' to modify. Be careful with the < and > signs. | ||
| + | You may include any references to papers as in: the use of JSmol in Proteopedia <ref>DOI 10.1002/ijch.201300024</ref> or to the article describing Jmol <ref>PMID:21638687</ref> to the rescue. | ||
| + | == Vitamin K Epoxide == | ||
| + | |||
| + | [[Image:Vitamin K epoxide.jpg|500 px|right|thumb|Figure 1. Vitamin K Epoxide structure]] | ||
| + | |||
| + | As mentioned above, Vitamin K epoxide is a part of the Vitamin K cycle, necessary for blood coagulation. In the cycle, Vitamin K epoxide reductase (VKOR) reduces Vitamin K epoxide to quinone, or the active form of Vitamin K. What is occurring is VKOR donated electrons to Vitamin K epoxide, and those electrons come from the S-H of one of the cysteine pairs discussed above. The one cysteine pair has to be reduced for the transfer of electrons to the substrate can occur. | ||
| + | |||
| + | |||
| + | === Binding === | ||
| + | To start, VKOR is in its open conformation. The Vitamin K epoxide enters. The oxygens of the ketones bind to <scene name='90/904322/Tyr_asn_binding/1'>Asn80 and Tyr139</scene>. With Vitamin K epoxide in its place, the conformation of VKOR is partially oxidized in regards to the cysteine pairs, which overall leads to the reduction of the substrate. A disulfide bond forms between Cys51 and Cys132, resulting in the closed conformation. This leaves the sulfur on Cys43 and the sulfur on Cys135 protonated. The available hydrogens on these cysteines are utilized in reducing the epoxide. First, the sulfur on Cys51 and Cys43 form a new bond. The hydrogen from Cys43 binds to the oxygen in the epoxide. The sulfur on Cys132 and the sulfur on Cys15 then form a new disulfide bond. The hydrogen that was present on Cys135 forms a new bond with the oxygen of the epoxide. With these cysteine pairs formed, VKOR is left in an open conformation. The end products are the Vitamin K/quinone and water. | ||
| + | |||
| + | |||
| + | |||
| + | == Warfarin == | ||
| + | |||
| + | |||
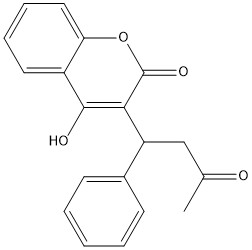
| + | [[Image:warfarin.jpg|400 px|right|thumb|Figure 1. Warfarin]] | ||
| + | |||
| + | |||
| + | |||
| + | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
| + | |||
| + | </StructureSection> | ||
| + | == References == | ||
| + | <references/> | ||
== Warfarin == | == Warfarin == | ||
Revision as of 20:19, 22 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Contents |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
Warfarin
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>