Sandbox Reserved 1716
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
<ref name="Ransey">PMID:28504306</ref> | <ref name="Ransey">PMID:28504306</ref> | ||
===Transmembrane Helices=== | ===Transmembrane Helices=== | ||
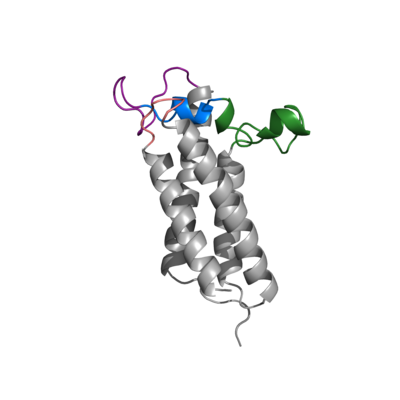
| + | The Transmembrane helices are named Transmembrane Helix 1, Transmembrane Helix 2, Transmembrane Helix 3, and Transmembrane Helix 4. The residues on Transmembrane Helix 2 (TM2) and Transmembrane Helix 4 (TM4) are significant for the binding of Vitamin K to the hydrophobic pocket of the enzyme. Asparagine 222 (make sure this is right) on TM2 and Tyrosine 281(make sure this is right) hydrogen bond to Vitamin K Epoxide, in order to hold it in place so that it may be reduced. Cysteine residues from the cap domain will donate their electrons to Vitamin K Epoxide to open the epoxide ring, and reform Vitamin K Quinone. | ||
| + | |||
===Cap Domain=== | ===Cap Domain=== | ||
Revision as of 15:50, 24 March 2022
Vitamin K Epoxide Reductase
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677