Sandbox Reserved 1713
From Proteopedia
| Line 13: | Line 13: | ||
[[Image:Proteo_ALK-ALKAL_Monomer_White.png|400 px|right|thumb|Figure 2]] | [[Image:Proteo_ALK-ALKAL_Monomer_White.png|400 px|right|thumb|Figure 2]] | ||
| - | <scene name='90/904318/Binding_surface_with_residues/3'>interacting residues of ALK and ALKAL</scene> | ||
<scene name='90/904318/Alkalbindingsurfacewmembrane/1'>Binding surface of ALKAL with the membrane</scene> | <scene name='90/904318/Alkalbindingsurfacewmembrane/1'>Binding surface of ALKAL with the membrane</scene> | ||
<scene name='90/904318/Alkal1membraneinteraction/2'>ALKAL's residues that interact with membrane</scene> | <scene name='90/904318/Alkal1membraneinteraction/2'>ALKAL's residues that interact with membrane</scene> | ||
===Conformational Change=== | ===Conformational Change=== | ||
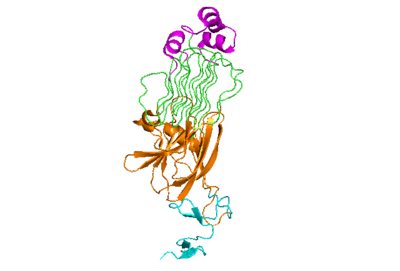
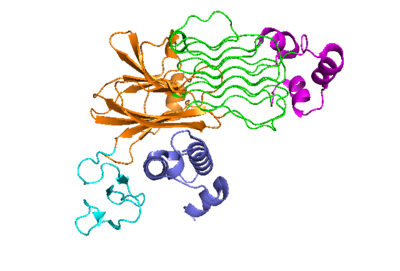
| - | The anaplastic lymphoma kinase activating ligand (ALKAL) binds to the <scene name='90/904318/Alk-alkal_binding_surface/2'>binding surface</scene> on the ALK at the TNFL domain. This induces a conformational change which allows for the PXL and the GlyR domains to hinge forward.<ref>DOI: 10.1038/s41586-021-04140-8</ref> (Figure 2) ALK's TNFL has <scene name='90/904318/Binding_surface_with_residues/3'>residues</scene> E978, E974, E859, and Y966 that form salt bridges with R123, R133, R136, R140, and R117 on ALKAL that allow for activation, leading to dimerization of two ALK-ALKAL monomers. | + | The anaplastic lymphoma kinase activating ligand (ALKAL) binds to the <scene name='90/904318/Alk-alkal_binding_surface/2'>binding surface</scene> on the ALK at the TNFL domain. This induces a conformational change which allows for the PXL and the GlyR domains to hinge forward.<ref>DOI: 10.1038/s41586-021-04140-8</ref> (Figure 2) ALK's TNFL has <scene name='90/904318/Binding_surface_with_residues/3'>residues</scene> E978, E974, E859, and Y966 that form salt bridges with R123, R133, R136, R140, and R117 on ALKAL that allow for activation, leading to <scene name='90/904317/Dimer_full_colored/3'>dimerization</scene> of two ALK-ALKAL monomers. |
===Movement across the membrane=== | ===Movement across the membrane=== | ||
The negatively charged phosphate groups on the cell membrane interact with the highly conserved positively charged residues on ALKAL ligand that face the membrane, which stabilizes the ligand to bind to ALK even better. <ref>DOI: 10.1038/s41586-021-04140-8</ref> | The negatively charged phosphate groups on the cell membrane interact with the highly conserved positively charged residues on ALKAL ligand that face the membrane, which stabilizes the ligand to bind to ALK even better. <ref>DOI: 10.1038/s41586-021-04140-8</ref> | ||
Revision as of 03:52, 28 March 2022
| This Sandbox is Reserved from February 28 through September 1, 2022 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1700 through Sandbox Reserved 1729. |
To get started:
More help: Help:Editing |
Anaplastic Lymphoma Kinase
Background
The anaplastic lymphoma kinase was first discovered in 1994 as a tyrosine kinase in a type of lymphoma cancer cell. ALK is a specific type of RTK which plays a huge role in transmembrane signaling and communication within the cell. ALK is commonly expressed in the development of the nervous system. Anaplastic Lymphoma Kinase receptor is a membrane-bound tyrosine kinase. The ALK transfers a phosphate group from ATP to a tyrosine residue on an enzyme which activates a signaling cascade, and ALK becomes activated when a ligand called ALKAL binds to the binding surface on an extracellular domain of ALK. ALK is an integral membrane protein. Abnormal forms of ALK are closely related to the formation of several cancers.
| |||||||||||
References
- ↑ Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ Jr, Shi X, Folta-Stogniew E, Gunel M, Alvarado D, Schlessinger J. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015 Jan 20;8(360):ra6. doi: 10.1126/scisignal.2005916. PMID:25605972 doi:http://dx.doi.org/10.1126/scisignal.2005916
- ↑ Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ Reshetnyak AV, Rossi P, Myasnikov AG, Sowaileh M, Mohanty J, Nourse A, Miller DJ, Lax I, Schlessinger J, Kalodimos CG. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature. 2021 Dec;600(7887):153-157. doi: 10.1038/s41586-021-04140-8. Epub 2021, Nov 24. PMID:34819673 doi:http://dx.doi.org/10.1038/s41586-021-04140-8
- ↑ De Munck S, Provost M, Kurikawa M, Omori I, Mukohyama J, Felix J, Bloch Y, Abdel-Wahab O, Bazan JF, Yoshimi A, Savvides SN. Structural basis of cytokine-mediated activation of ALK family receptors. Nature. 2021 Oct 13. pii: 10.1038/s41586-021-03959-5. doi:, 10.1038/s41586-021-03959-5. PMID:34646012 doi:http://dx.doi.org/10.1038/s41586-021-03959-5
- ↑ Li T, Stayrook SE, Tsutsui Y, Zhang J, Wang Y, Li H, Proffitt A, Krimmer SG, Ahmed M, Belliveau O, Walker IX, Mudumbi KC, Suzuki Y, Lax I, Alvarado D, Lemmon MA, Schlessinger J, Klein DE. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature. 2021 Dec;600(7887):148-152. doi: 10.1038/s41586-021-04141-7. Epub 2021, Nov 24. PMID:34819665 doi:http://dx.doi.org/10.1038/s41586-021-04141-7