Sandbox Reserved 1715
From Proteopedia
(Difference between revisions)
| Line 49: | Line 49: | ||
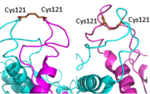
[[Image: Active vs inactive vft.png|150px|left|thumb|Figure 1. Cysteine 121 positioning in the inactive (left) vs the active (right) mGlu conformation ]] | [[Image: Active vs inactive vft.png|150px|left|thumb|Figure 1. Cysteine 121 positioning in the inactive (left) vs the active (right) mGlu conformation ]] | ||
==== Domains ==== | ==== Domains ==== | ||
| - | '''VFT''': The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a disulfide bond which is shifted down in the inactive form | + | '''VFT''': The extracellular location in which the two glutamate agonists bind is known as the VFT. This domain includes a disulfide bond which is shifted down in the inactive form and undergoes an upward movement upon glutamate binding which stabilizes the active site (Figure 1). |
'''CRD''': The portion of the protomer that connects the VFT with the TMD is known as the CRD. As the linking segment of the protein, is critical in transmitting the conformational change caused by the binding of glutamate to the TMD. The change resulting from the binding of glutamate in the VFT brings the cysteine-rich domain together to alter the configuration of the TMD through its interaction with the extracellular loop 2 (ECL2). This process is mediated by the hydrophobic effect due to the nature of the amino acids at the apex of the CRD.( 3D IMAGE) | '''CRD''': The portion of the protomer that connects the VFT with the TMD is known as the CRD. As the linking segment of the protein, is critical in transmitting the conformational change caused by the binding of glutamate to the TMD. The change resulting from the binding of glutamate in the VFT brings the cysteine-rich domain together to alter the configuration of the TMD through its interaction with the extracellular loop 2 (ECL2). This process is mediated by the hydrophobic effect due to the nature of the amino acids at the apex of the CRD.( 3D IMAGE) | ||
| Line 57: | Line 57: | ||
== Conformational Changes == | == Conformational Changes == | ||
| - | '''1.''' mGlu starts in an <scene name='90/904320/Inactive_mglu/2'>inactive homodimeric form</scene>. In this conformation, the receptor is considered open with an inter-lobe angle of 44 degrees.The structure has two free binding sites in the VFT, the CRDs are separated, and the TMD is not interacting with a G protein. [[Image: Protein Interaction with G Protein.png|300 px|right|thumb|Figure | + | '''1.''' mGlu starts in an <scene name='90/904320/Inactive_mglu/2'>inactive homodimeric form</scene>. In this conformation, the receptor is considered open with an inter-lobe angle of 44 degrees.The structure has two free binding sites in the VFT, the CRDs are separated, and the TMD is not interacting with a G protein. [[Image: Protein Interaction with G Protein.png|300 px|right|thumb|Figure 2. The interaction between an active mGlu and a G-protein ]] |
'''2.''' In the intermediate activation state (known as the open-closed conformation), one glutamate is bound in one binding pocket of VFT. This <scene name='90/904320/Mglu_binding/4'>glutamate bound state</scene> is still considered inactive as the receptor has not changed the conformations in the CRD and thus the TMD. With the same asymmetric transmembrane helices formation, a TM3-TM4 interface is still present and mGlu cannot interact with a G protein. (IMAGE) | '''2.''' In the intermediate activation state (known as the open-closed conformation), one glutamate is bound in one binding pocket of VFT. This <scene name='90/904320/Mglu_binding/4'>glutamate bound state</scene> is still considered inactive as the receptor has not changed the conformations in the CRD and thus the TMD. With the same asymmetric transmembrane helices formation, a TM3-TM4 interface is still present and mGlu cannot interact with a G protein. (IMAGE) | ||
| - | '''3.''' A second glutamate binds to the other <scene name='90/904320/Active_site_interactions/3'>binding pocket</scene> of the VFT. Mediated by L639, F643, N735, W773, and F776, a positive allosteric modulator (PAM) also binds within the seven TMD helices of the alpha chain. This closed conformation with an inter-lobe domain of 25 degrees is considered the active conformation. The binding of these ligands allows the CRD to compact and come together. This transformation causes the TMD to form another asymmetric conformation with a TM6-TM6 interface between the chains. | + | '''3.''' A second glutamate binds to the other <scene name='90/904320/Active_site_interactions/3'>binding pocket</scene> of the VFT. Mediated by L639, F643, N735, W773, and F776, a positive allosteric modulator (PAM) also binds within the seven TMD helices of the alpha chain. This closed conformation with an inter-lobe domain of 25 degrees is considered the <scene name='90/904319/active conformation/1'>PAM</scene>. The binding of these ligands allows the CRD to compact and come together. This transformation causes the TMD to form another asymmetric conformation with a TM6-TM6 interface between the chains. |
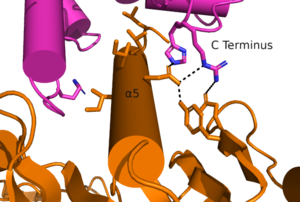
'''4.''' The change in the arrangement of the helices allows for intracellular loop 2 (ICL2) and the C-terminus to be properly ordered to interact with a G protein. While hydrogen bonding is present, this coupling is primarily driven by the hydrophobic interactions in the interface with the ɑ5 helix of the G protein.. This coupling can only occur in the presence of a PAM as the pocket in which the coupling occurs would be completely closed in its absence. | '''4.''' The change in the arrangement of the helices allows for intracellular loop 2 (ICL2) and the C-terminus to be properly ordered to interact with a G protein. While hydrogen bonding is present, this coupling is primarily driven by the hydrophobic interactions in the interface with the ɑ5 helix of the G protein.. This coupling can only occur in the presence of a PAM as the pocket in which the coupling occurs would be completely closed in its absence. | ||
Revision as of 05:48, 28 March 2022
Metabotropic Glutamate Receptor
| |||||||||||
References
- ↑ 1.0 1.1 Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ 2.0 2.1 Seven AB, Barros-Alvarez X, de Lapeyriere M, Papasergi-Scott MM, Robertson MJ, Zhang C, Nwokonko RM, Gao Y, Meyerowitz JG, Rocher JP, Schelshorn D, Kobilka BK, Mathiesen JM, Skiniotis G. G-protein activation by a metabotropic glutamate receptor. Nature. 2021 Jun 30. pii: 10.1038/s41586-021-03680-3. doi:, 10.1038/s41586-021-03680-3. PMID:34194039 doi:http://dx.doi.org/10.1038/s41586-021-03680-3
Student Contributors
- Courtney Vennekotter
- Cade Chezem