We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1709
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Structural Highlights== | == Structural Highlights== | ||
=== Active Site === | === Active Site === | ||
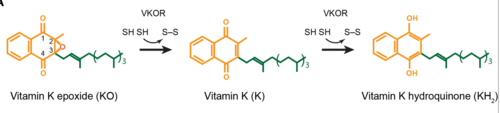
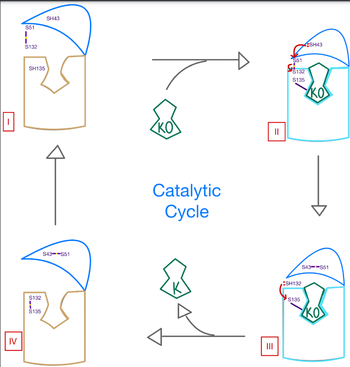
| + | Within the four transmembrane helices lies the active site. The active site is comprised of a hydrophobic pocket containing two hydrophilic residues, A80 and Y139, that interact with substrates and ligands alike. The hydrophobic pocket provides specificity to the region while the hydrophilic residues have potential to hydrogen bond, allowing recognition and increasing specificity as well. Slightly above the active site is a crucial disulfide bridge that provides stabilization when a substrate is bound. This bridge occurs between C132 and C135, recurrent residues that continually aid in VKOR function. | ||
| + | |||
The <scene name='90/906893/Active_site/2'>active site</scene> plays a vital role in binding of any substrate or ligand to the VKOR. Upon binding, the VKOR will transition into a <scene name='90/906893/Closed_conformation/4'>closed conformation</scene> that will allow its catalytic mechanism to commence. | The <scene name='90/906893/Active_site/2'>active site</scene> plays a vital role in binding of any substrate or ligand to the VKOR. Upon binding, the VKOR will transition into a <scene name='90/906893/Closed_conformation/4'>closed conformation</scene> that will allow its catalytic mechanism to commence. | ||
Revision as of 22:17, 28 March 2022
| |||||||||||
References
1. Li, Weikai et al. “Structure of a bacterial homologue of vitamin K epoxide reductase.” Nature vol. 463,7280 (2010): 507-12. doi:10.1038/nature08720.
2. Liu S, Li S, Shen G, Sukumar N, Krezel AM, Li W. Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science. 2021 Jan 1;371(6524):eabc5667. doi: 10.1126/science.abc5667. Epub 2020 Nov 5. PMID: 33154105; PMCID: PMC7946407.
3. “Warfarin.” Wikipedia, Wikimedia Foundation, 10 Feb. 2022, https://en.wikipedia.org/wiki/Warfarin.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644