Sandbox Reserved 1711
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
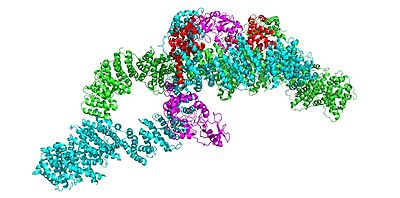
=== Domains === | === Domains === | ||
| - | The <scene name='90/904316/Grd_domains/ | + | The <scene name='90/904316/Grd_domains/2'>GRD</scene> of Neurofibromin, specifically the arginine finger (R1276), binds to the Ras + GTP complex. |
=== Key Players === | === Key Players === | ||
An <scene name='90/905640/Arg_finger/1'>R1276</scene> (R1276) <scene name='90/904316/Arg_finger/1'>R1276</scene> present in the GRD is critical for Ras binding and is only accessible when the GRD and Sec14-PH domains are rotated in such a way that there is no steric hindrance from the surrounding dimer chains. The Closed conformation is stabilized by a <scene name='90/904316/Closed_triade/1'>triade</scene> of residues that are coordinated with transition metal-binding sites with zinc. Here, the GRD and Sec14-PH domains are oriented in a way that the H1558 and H1576 are able to interact with C1032 and form a transition binding-site with zinc. This binding site stabilizes the closed conformation and prevents Ras from associating with the GRD based on the location of the GRD in relation to the rest of the protein. | An <scene name='90/905640/Arg_finger/1'>R1276</scene> (R1276) <scene name='90/904316/Arg_finger/1'>R1276</scene> present in the GRD is critical for Ras binding and is only accessible when the GRD and Sec14-PH domains are rotated in such a way that there is no steric hindrance from the surrounding dimer chains. The Closed conformation is stabilized by a <scene name='90/904316/Closed_triade/1'>triade</scene> of residues that are coordinated with transition metal-binding sites with zinc. Here, the GRD and Sec14-PH domains are oriented in a way that the H1558 and H1576 are able to interact with C1032 and form a transition binding-site with zinc. This binding site stabilizes the closed conformation and prevents Ras from associating with the GRD based on the location of the GRD in relation to the rest of the protein. | ||
Revision as of 19:00, 29 March 2022
Neurofibromin
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Hannah Luchinski