We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='' scene='90/904325/Introductory_image_full/1' size='340' side='right' caption='Neurofibromin'> | <StructureSection load='' scene='90/904325/Introductory_image_full/1' size='340' side='right' caption='Neurofibromin'> | ||
== Introduction == | == Introduction == | ||
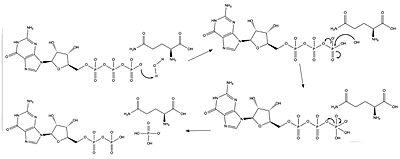
| - | Neurofibromin (NF) is a cytoplasmic protein encoded by the ''NF1'' gene located on chromosome 17 <ref name= ''Bergoug''>PMID:33121128</ref>. NF suppresses the [https://proteopedia.org/wiki/index.php/GTPase_HRas "Ras"] oncogene | + | Neurofibromin (NF) is a cytoplasmic protein encoded by the ''NF1'' gene located on chromosome 17 <ref name= ''Bergoug''>PMID:33121128</ref>. The neurofibromin protein in encoded by over 350 kb of DNA and contains 62 exons, 58 of whihc are constitutive<ref name= ''Trovó-Marqui''>PMID:16813595</ref>. NF suppresses the [https://proteopedia.org/wiki/index.php/GTPase_HRas "Ras"] oncogene by increasing the rate of hydrolysis from <scene name='90/904325/Ras_full_structure/2'>Ras-GTP</scene> (active) to Ras-GDP (inactive)<ref name= ''Hall''>PMID:12213964</ref>. Increasing the rate of Ras inactivation decreases cell proliferation linked to cancer<ref name= ''Cimino''>PMID:29478615</ref>. |
== Structure == | == Structure == | ||
| Line 29: | Line 29: | ||
== Disease Relevance == | == Disease Relevance == | ||
| - | Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1 (NF1). This condition drives several forms of human cancers by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people, and the NF gene itself has the highest mutation rate of any known human gene, adding to its prevalence<ref name= ''Lupton''>PMID:34887559</ref>. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems<ref name= ''Cimino''>PMID:29478615</ref>. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein<ref name= ''Ly''>PMID:31582003</ref>. | + | Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1 (NF1). This condition drives several forms of human cancers by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people, and the NF gene itself has the highest mutation rate of any known human gene, adding to its prevalence<ref name= ''Lupton''>PMID:34887559</ref>. Furthermore, these mutations consist heavily of ''de novo'' mutations<ref name= ''Abramowicz''>PMID:25182393</ref>. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems<ref name= ''Cimino''>PMID:29478615</ref>. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein<ref name= ''Ly''>PMID:31582003</ref>. |
Revision as of 19:18, 7 April 2022
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12138-42. Epub 2002 Sep 4. PMID:12213964 doi:http://dx.doi.org/10.1073/pnas.192453199
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Frech M, Darden TA, Pedersen LG, Foley CK, Charifson PS, Anderson MW, Wittinghofer A. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 1994 Mar 22;33(11):3237-44. doi: 10.1021/bi00177a014. PMID:8136358 doi:http://dx.doi.org/10.1021/bi00177a014
- ↑ Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, Raught B, Zhang ZY, Zadeh G, Ohh M. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat Commun. 2015 Nov 30;6:8859. doi: 10.1038/ncomms9859. PMID:26617336 doi:http://dx.doi.org/10.1038/ncomms9859
- ↑ Scheffzek K, Shivalingaiah G. Ras-Specific GTPase-Activating Proteins-Structures, Mechanisms, and Interactions. Cold Spring Harb Perspect Med. 2019 Mar 1;9(3). pii: cshperspect.a031500. doi:, 10.1101/cshperspect.a031500. PMID:30104198 doi:http://dx.doi.org/10.1101/cshperspect.a031500
- ↑ Prive GG, Milburn MV, Tong L, de Vos AM, Yamaizumi Z, Nishimura S, Kim SH. X-ray crystal structures of transforming p21 ras mutants suggest a transition-state stabilization mechanism for GTP hydrolysis. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3649-53. doi: 10.1073/pnas.89.8.3649. PMID:1565661 doi:http://dx.doi.org/10.1073/pnas.89.8.3649
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Abramowicz A, Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1gene as a cause of disease. Dev Period Med. 2014 Jul-Sep;18(3):297-306. PMID:25182393
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky