We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Neurofibromin
From Proteopedia
(Difference between revisions)
| Line 22: | Line 22: | ||
==RAS Complex== | ==RAS Complex== | ||
| - | + | ||
| - | + | ||
| - | + | ||
== Disease Relevance == | == Disease Relevance == | ||
| - | Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1 | + | Mutations to the neurofibromin protein are implicated in the progression of Neurofibromatosis type 1. Neurofibromatosis type 1 is a condition where cancer develops by inactivating the Ras suppression effects of NF, allowing Ras to behave as an oncogene. Neurofibromatosis type 1 is an autosomal dominant disorder that affects 1 in 3,000 people. The ''NF'' gene has the highest mutation rate of any human gene, adding to the prevalence of cancers related to neurofibromin mutations<ref name= ''Lupton''>PMID:34887559</ref>. Furthermore, these mutations consist heavily of ''de novo'' mutations<ref name= ''Abramowicz''>PMID:25182393</ref>. NF1 primarily causes tumors in the central and peripheral nervous systems, but often has a multisystem expression including tumors in the dermatologic, cardiovascular, gastrointestinal, and orthopedic systems<ref name= ''Cimino''>PMID:29478615</ref>. The wide range of presentations is consistent with the multiplicity of mutations observed in the causative protein<ref name= ''Ly''>PMID:31582003</ref>. This multiplicity derives from the immense size and homo dimeric nature of the neurofibromin protein that allows for otherwise innocuous mutations to wreak havoc on the conformations of the protein as well as its ability to bind to the Ras protein as positioning of NF is highly important. |
| + | |||
| + | ===Downstream Effects=== | ||
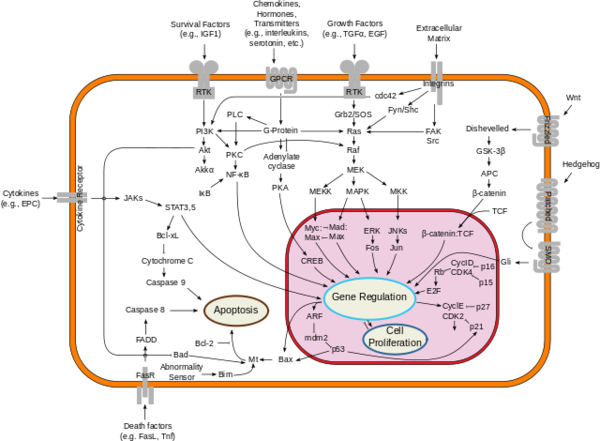
| + | [[Image:Signal_transduction_pathways.png|600 px|right|thumb|Figure 2: Downstream proliferation of Ras signals have a variety of impacts, including gene regulation, cell proliferation, and cell growth; By cybertory - This file was derived from: Signal transduction v1.png, CC BY-SA 3.0, https://commons.wikimedia.org/w/index.php?curid=12081090]] | ||
| + | Misregulated [http://https://proteopedia.org/wiki/index.php/GTPase_HRas Ras activity] can lead to uncontrolled signaling in many different cell signaling pathways. Figure 2 provides an overview of the pathways that are connected to Ras, such as the MEK pathway. | ||
Revision as of 19:33, 7 April 2022
| |||||||||||
References
- ↑ Bergoug M, Doudeau M, Godin F, Mosrin C, Vallee B, Benedetti H. Neurofibromin Structure, Functions and Regulation. Cells. 2020 Oct 27;9(11). pii: cells9112365. doi: 10.3390/cells9112365. PMID:33121128 doi:http://dx.doi.org/10.3390/cells9112365
- ↑ Trovo-Marqui AB, Tajara EH. Neurofibromin: a general outlook. Clin Genet. 2006 Jul;70(1):1-13. doi: 10.1111/j.1399-0004.2006.00639.x. PMID:16813595 doi:http://dx.doi.org/10.1111/j.1399-0004.2006.00639.x
- ↑ Hall BE, Bar-Sagi D, Nassar N. The structural basis for the transition from Ras-GTP to Ras-GDP. Proc Natl Acad Sci U S A. 2002 Sep 17;99(19):12138-42. Epub 2002 Sep 4. PMID:12213964 doi:http://dx.doi.org/10.1073/pnas.192453199
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Frech M, Darden TA, Pedersen LG, Foley CK, Charifson PS, Anderson MW, Wittinghofer A. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: an experimental and theoretical study. Biochemistry. 1994 Mar 22;33(11):3237-44. doi: 10.1021/bi00177a014. PMID:8136358 doi:http://dx.doi.org/10.1021/bi00177a014
- ↑ Bunda S, Burrell K, Heir P, Zeng L, Alamsahebpour A, Kano Y, Raught B, Zhang ZY, Zadeh G, Ohh M. Inhibition of SHP2-mediated dephosphorylation of Ras suppresses oncogenesis. Nat Commun. 2015 Nov 30;6:8859. doi: 10.1038/ncomms9859. PMID:26617336 doi:http://dx.doi.org/10.1038/ncomms9859
- ↑ Lupton CJ, Bayly-Jones C, D'Andrea L, Huang C, Schittenhelm RB, Venugopal H, Whisstock JC, Halls ML, Ellisdon AM. The cryo-EM structure of the human neurofibromin dimer reveals the molecular basis for neurofibromatosis type 1. Nat Struct Mol Biol. 2021 Dec;28(12):982-988. doi: 10.1038/s41594-021-00687-2., Epub 2021 Dec 9. PMID:34887559 doi:http://dx.doi.org/10.1038/s41594-021-00687-2
- ↑ Abramowicz A, Gos M. Neurofibromin in neurofibromatosis type 1 - mutations in NF1gene as a cause of disease. Dev Period Med. 2014 Jul-Sep;18(3):297-306. PMID:25182393
- ↑ Cimino PJ, Gutmann DH. Neurofibromatosis type 1. Handb Clin Neurol. 2018;148:799-811. doi: 10.1016/B978-0-444-64076-5.00051-X. PMID:29478615 doi:http://dx.doi.org/10.1016/B978-0-444-64076-5.00051-X
- ↑ Ly KI, Blakeley JO. The Diagnosis and Management of Neurofibromatosis Type 1. Med Clin North Am. 2019 Nov;103(6):1035-1054. doi: 10.1016/j.mcna.2019.07.004. PMID:31582003 doi:http://dx.doi.org/10.1016/j.mcna.2019.07.004
Proteopedia Page Contributors and Editors (what is this?)
Jordyn K. Lenard, Ryan D. Adkins, Michal Harel, OCA, Jaime Prilusky