We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1703
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
===Overall Structure=== | ===Overall Structure=== | ||
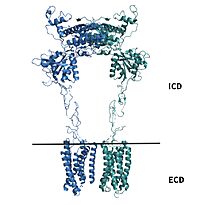
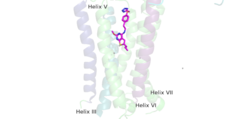
| - | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM] studies of mGlu2 have yielded adequate structural maps of mGlu2 in various activation states. These maps provided clearer understanding of the conformational changes between the inactive and active states of mGlu2<ref name="Lin" />. The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Vft/1'>Venus FlyTrap Domain(VFT)</scene>, followed by a <scene name='90/904307/Crds/1'>Cysteine Rich Domain</scene> linker to the Transmembrane Domain that contains <scene name='90/904307/Inactive_7tm/1'>7 alpha helices (7TM)</scene> on both the <scene name='90/904308/Alphaandbetachain/1'>alpha and beta</scene> chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. | ||
[[Image:Domains of mGlu2.jpg|200 px|left thumb|'''Figure 2.'''Show the regions of mGlu2.]] | [[Image:Domains of mGlu2.jpg|200 px|left thumb|'''Figure 2.'''Show the regions of mGlu2.]] | ||
| + | [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo-EM] studies of mGlu2 have yielded adequate structural maps of mGlu2 in various activation states. These maps provided clearer understanding of the conformational changes between the inactive and active states of mGlu2<ref name="Lin" />. The overall <scene name='90/904307/Inactive_structure/1'>structure</scene> of the mGlu2 is composed of 3 main parts: a ligand binding <scene name='90/904307/Vft/1'>Venus FlyTrap Domain(VFT)</scene>, followed by a <scene name='90/904307/Crds/1'>Cysteine Rich Domain</scene> linker to the Transmembrane Domain that contains <scene name='90/904307/Inactive_7tm/1'>7 alpha helices (7TM)</scene> on both the <scene name='90/904308/Alphaandbetachain/1'>alpha and beta</scene> chains that aid in the binding of the G-Protein. Class C CPCRs such as mGlu2, are activated by their ability to form dimers. | ||
| + | |||
mGlu2 is a [https://en.wikipedia.org/wiki/Protein_dimer homodimer]. Dimerization of mGlu2 is required to relay glutamate binding from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | mGlu2 is a [https://en.wikipedia.org/wiki/Protein_dimer homodimer]. Dimerization of mGlu2 is required to relay glutamate binding from the extracellular domain(ECD) to its transmembrane domain(TMD). The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | ||
| + | |||
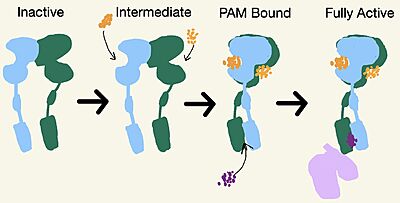
[[Image:Schematic of mGlu2.jpg|400 px|right|thumb|'''Figure 3.''' Demonstrates the conformational changes of mGlu2.]] | [[Image:Schematic of mGlu2.jpg|400 px|right|thumb|'''Figure 3.''' Demonstrates the conformational changes of mGlu2.]] | ||
Revision as of 18:06, 14 April 2022
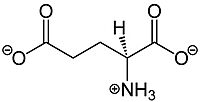
Metabotropic Glutamate Receptor 2
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Lin S, Han S, Cai X, Tan Q, Zhou K, Wang D, Wang X, Du J, Yi C, Chu X, Dai A, Zhou Y, Chen Y, Zhou Y, Liu H, Liu J, Yang D, Wang MW, Zhao Q, Wu B. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021 Jun;594(7864):583-588. doi: 10.1038/s41586-021-03495-2. Epub 2021, Jun 16. PMID:34135510 doi:http://dx.doi.org/10.1038/s41586-021-03495-2
- ↑ 2.0 2.1 Seven, Alpay B., et al. “G-Protein Activation by a Metabotropic Glutamate Receptor.” Nature News, Nature Publishing Group, 30 June 2021, https://www.nature.com/articles/s1586-021-03680-3
- ↑ Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.>
- ↑ 4.0 4.1 4.2 4.3 Muguruza C, Meana JJ, Callado LF. Group II Metabotropic Glutamate Receptors as Targets for Novel Antipsychotic Drugs. Front Pharmacol. 2016 May 20;7:130. doi: 10.3389/fphar.2016.00130. eCollection, 2016. PMID:27242534 doi:http://dx.doi.org/10.3389/fphar.2016.00130
- ↑ Ellaithy A, Younkin J, Gonzalez-Maeso J, Logothetis DE. Positive allosteric modulators of metabotropic glutamate 2 receptors in schizophrenia treatment. Trends Neurosci. 2015 Aug;38(8):506-16. doi: 10.1016/j.tins.2015.06.002. Epub, 2015 Jul 4. PMID:26148747 doi:http://dx.doi.org/10.1016/j.tins.2015.06.002
Student Contributors
Frannie Brewer and Ashley Wilkinson