Sandbox Reserved 1703
From Proteopedia
(Difference between revisions)
| Line 15: | Line 15: | ||
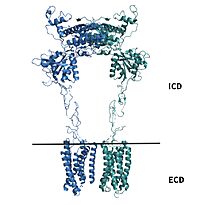
mGlu2 is a [https://en.wikipedia.org/wiki/Protein_dimer homodimer]. Dimerization of mGlu2 is required to relay glutamate binding from the ECD to its transmembrane domain TMD. The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | mGlu2 is a [https://en.wikipedia.org/wiki/Protein_dimer homodimer]. Dimerization of mGlu2 is required to relay glutamate binding from the ECD to its transmembrane domain TMD. The homodimer of mGlu2 contains an alpha chain and a beta chain. Occupation of both ECDs with the agonist, glutamate, is necessary for a fully active mGlu2<ref name="Du">Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.></ref>. However, only one chain in the dimer is responsible for activation of the G-protein, this suggests an asymmetrical signal transduction mechanism for mGlu2<ref name="Lin"/>. | ||
| - | |||
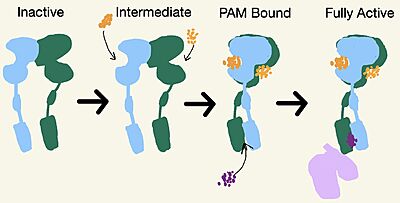
| - | [[Image:Schematic of mGlu2.jpg|400 px|left|thumb|'''Figure 3.''' Demonstrates the conformational changes of mGlu2.]] | ||
===Inactive State=== | ===Inactive State=== | ||
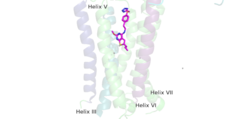
A few hallmarks of the inactive structure of mGlu2 are the <scene name='90/904307/Better_inactive_structure/3'>Venus Fly Trap Domain (VFT)</scene> in the open conformation, well separated <scene name='90/904307/Better_inactive_structure/2'>Cysteine Rich Domain</scene>, and distinct orientation of the 7 Transmembrane Domains (7TM). The most critical component of the inactive form is the <scene name='90/904307/Tmd_helices/4'>asymmetric TM3-TM4 interface</scene> formed by the 7 α-helices in the α and β chains of the 7TM. The inactive structure of mGlu2 is mediated mainly by helices 3 and 4 on both the α and β chains of the dimer through hydrophobic interactions. These <scene name='90/904307/Tm3-tm4_hydrophobic/2'>hydrophobic interactions</scene> between both transmembrane helices stabilize inactive conformation of mGlu2<ref name="Lin"/>. Key hydrophobic interactions are between A630 V699 on the helix | A few hallmarks of the inactive structure of mGlu2 are the <scene name='90/904307/Better_inactive_structure/3'>Venus Fly Trap Domain (VFT)</scene> in the open conformation, well separated <scene name='90/904307/Better_inactive_structure/2'>Cysteine Rich Domain</scene>, and distinct orientation of the 7 Transmembrane Domains (7TM). The most critical component of the inactive form is the <scene name='90/904307/Tmd_helices/4'>asymmetric TM3-TM4 interface</scene> formed by the 7 α-helices in the α and β chains of the 7TM. The inactive structure of mGlu2 is mediated mainly by helices 3 and 4 on both the α and β chains of the dimer through hydrophobic interactions. These <scene name='90/904307/Tm3-tm4_hydrophobic/2'>hydrophobic interactions</scene> between both transmembrane helices stabilize inactive conformation of mGlu2<ref name="Lin"/>. Key hydrophobic interactions are between A630 V699 on the helix | ||
| + | [[Image:Schematic of mGlu2.jpg|400 px|left|thumb|'''Figure 3.''' Demonstrates the conformational changes of mGlu2.]] | ||
===Intermediate Form=== | ===Intermediate Form=== | ||
Revision as of 19:59, 18 April 2022
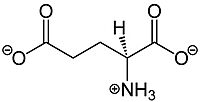
Metabotropic Glutamate Receptor 2
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 1.8 Lin S, Han S, Cai X, Tan Q, Zhou K, Wang D, Wang X, Du J, Yi C, Chu X, Dai A, Zhou Y, Chen Y, Zhou Y, Liu H, Liu J, Yang D, Wang MW, Zhao Q, Wu B. Structures of Gi-bound metabotropic glutamate receptors mGlu2 and mGlu4. Nature. 2021 Jun;594(7864):583-588. doi: 10.1038/s41586-021-03495-2. Epub 2021, Jun 16. PMID:34135510 doi:http://dx.doi.org/10.1038/s41586-021-03495-2
- ↑ 2.0 2.1 Seven, Alpay B., et al. “G-Protein Activation by a Metabotropic Glutamate Receptor.” Nature News, Nature Publishing Group, 30 June 2021, https://www.nature.com/articles/s1586-021-03680-3
- ↑ Du, Juan, et al. “Structures of Human mglu2 and mglu7 Homo- and Heterodimers.” Nature News, Nature Publishing Group, 16 June 2021, https://www.nature.com/articles/s41586-021-03641-w.>
- ↑ \“Schizophrenia.” National Institute of Mental Health, U.S. Department of Health and Human Services, https://www.nimh.nih.gov/health/topics/schizophrenia
- ↑ 5.0 5.1 Ellaithy A, Younkin J, Gonzalez-Maeso J, Logothetis DE. Positive allosteric modulators of metabotropic glutamate 2 receptors in schizophrenia treatment. Trends Neurosci. 2015 Aug;38(8):506-16. doi: 10.1016/j.tins.2015.06.002. Epub, 2015 Jul 4. PMID:26148747 doi:http://dx.doi.org/10.1016/j.tins.2015.06.002
- ↑ 6.0 6.1 6.2 6.3 Muguruza C, Meana JJ, Callado LF. Group II Metabotropic Glutamate Receptors as Targets for Novel Antipsychotic Drugs. Front Pharmacol. 2016 May 20;7:130. doi: 10.3389/fphar.2016.00130. eCollection, 2016. PMID:27242534 doi:http://dx.doi.org/10.3389/fphar.2016.00130
Student Contributors
Frannie Brewer and Ashley Wilkinson